Biomed Res Bull. 1(2):66-76.
doi: 10.34172/biomedrb.2023.13
Review Article
Application and Immunogenicity of Aptamers in Cancer Therapy
Arin Ghazizadeh 1  , Raha Amazadeh 2, Mahyar Ziyamolki 1, Amirhossein Olfati 1, Shalen Kumar 3, Tooba Gholikhani 1, 2, 4, *
, Raha Amazadeh 2, Mahyar Ziyamolki 1, Amirhossein Olfati 1, Shalen Kumar 3, Tooba Gholikhani 1, 2, 4, * 
Author information:
1Faculty of Pharmacy, Tabriz University of Medical Sciences, Tabriz, Iran
2Student Research Committee, Tabriz University of Medical Sciences, Tabriz, Iran
3School of Biological Sciences, Victoria University of Wellington, Kelburn parade, Wellington 6021, New Zealand 5 IQ Science Ltd, Wellington, New Zealand
4Nanora Pharmaceuticals, Tabriz, Iran
Abstract
Aptamers are short single-stranded nucleic acid (DNA or RNA) or peptide sequences with a unique three-dimensional structure that bind to their targets with high specificity and affinity. Aptamers’ small size, fast and cheap production process, low immunogenicity, and high stability have made them attractive molecules in some fields of research, especially in the design of new diagnostic and therapeutic pathways. This review article discusses aptamers’ applications and immunogenic properties in the pharmaceutical sciences.
Keywords: Applications, Aptamer, Immunogenicity, Pharmaceutical sciences
Copyright and License Information
© 2023 The Author(s).
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
The word aptamer comes from the Latin word “Aptus” which means inclusion. It is a single-stranded nucleic acid (DNA or RNA) or oligopeptide, specifically capable of binding to a high-affinity target.1 Aptamers have many targets, ranging from small molecules and macromolecules to cells (prokaryotic and eukaryotic).2 Certain three-dimensional (3D) spatial forms, such as stem-loops, hairpins, quadruplexes, triplexes, triple bonds, and the like, have been developed based on aptamers’ nucleic acid arrangements. Aptamers have a high affinity for folding target molecules that can specifically bind to their destinations. Substances such as metal ions (Hg2+ and Pb2+ and the like), small organic molecules (amino acids, adenosine tri-amino acids, antibiotics, vitamins, and drugs), macromolecules (e.g., peptides, proteins, growth factors, dyes, toxins, pathogens, and biological markers), and microorganisms (e.g., bacteria) can be detected and measured due to the high affinity of aptamers for a variety of analytes. This article discusses a group of aptamers named oligonucleotides.2 Over the last decade, fluorescent semiconductor quantum dot compounds of aptamers have become an effective substratum for imaging and treatment of different illnesses (especially cancer) in animal and laboratory models.3
Moreover, these conjugates have widespread potential to monitor the environment, diagnose diseases, and use bioassays. The study demonstrates recent discoveries of aptamer’s immunogenicity properties in a variety of treatment applications. The single-stranded DNA, known as an aptamer, can bind to target molecules and disable them. Several researchers have also developed a more efficient DNA system to diagnose and treat infectious and cancer diseases.4 Scientists have improved the existing technologies for the synthesis of a modified DNA molecule called an aptamer. DNA aptamers are highly useful for pharmaceutical purposes since they can specifically bind to body molecular targets such as proteins, viruses, bacteria, and cells.5 DNA aptamers bind to the target and prevent it from functioning since they are manufactured synthetically. DNA aptamers have become an excellent technology for the diagnosis and delivery of medicines. However, no DNA aptamer has yet been authorized for clinical use because it is neither bound well nor digested easily by enzymes. Research by Yang et al revealed that a synthesizer is a DNA aptamer that can connect strongly to target molecules and has long-lasting stability to overcome these challenges. The results are published in the Scientific Reports journal.6 To solve the problem of poor binding, the research team of Yang et al added an abnormal base to the DNA aptamer (which has four main bases). The increased fifth component significantly enhances the aptamer’s ability to attach to the target molecule (binding power increases 100-fold compared to conventional aptamer). Moreover, a small, unique part of DNA (called a mini-DNA clamp) is added to the DNA aptamer to avoid the simple digestion of the aptamer by enzymes.7 If the technology is commercialized effectively, DNA aptamers could also replace or supplement disease treatment antibodies. Antibodies such as aptamers are linked to specific targets but often lead to adverse immune reactions, whereas the extensive production process with high quality is not easy.8 Aptamers have unique properties, including being considerably smaller than antibodies and enzymes, making them superior to other detection molecules. The non-specific uptake of nucleic acids is usually weaker compared to proteins.9 DNA aptamers are commonly used for the design of a wide variety of pharmaceutical vaccines and medicines with high stability because of the aptamers’ excellent immunogenicity. The main limitation of RNA aptamers is ribonuclease degradation, which is inhibited by diethylpyrocarbonate. The aptamer affinity of its particular analyte is occasionally up to ten thousand times greater than that of a similar molecule. This process is highly similar to a binding process for antigens and antibodies; thus, aptamers are sometimes called nucleotide antibodies.10 Therefore, aptamers’ immunogenicity is extremely important today, especially in the vaccine industry and particularly in pharmacy. This study aims to evaluate the immunogenicity of aptamers in pharmaceutical sciences.11
Aptamer Structure
Aptamer oligonucleotides are single-stranded or short-stranded RNAs that consist of approximately 20-80 nucleotides (5-15 kDa). Aptamers (Figure 1) can be found in complex folding structures such as quadruplex, pseudoknot, bulge, stem-loop, and aptamer oligonucleotides. These complex structures allow aptamers to bind specifically to their targets; in fact, aptamers identify their goals by structural recognition.12
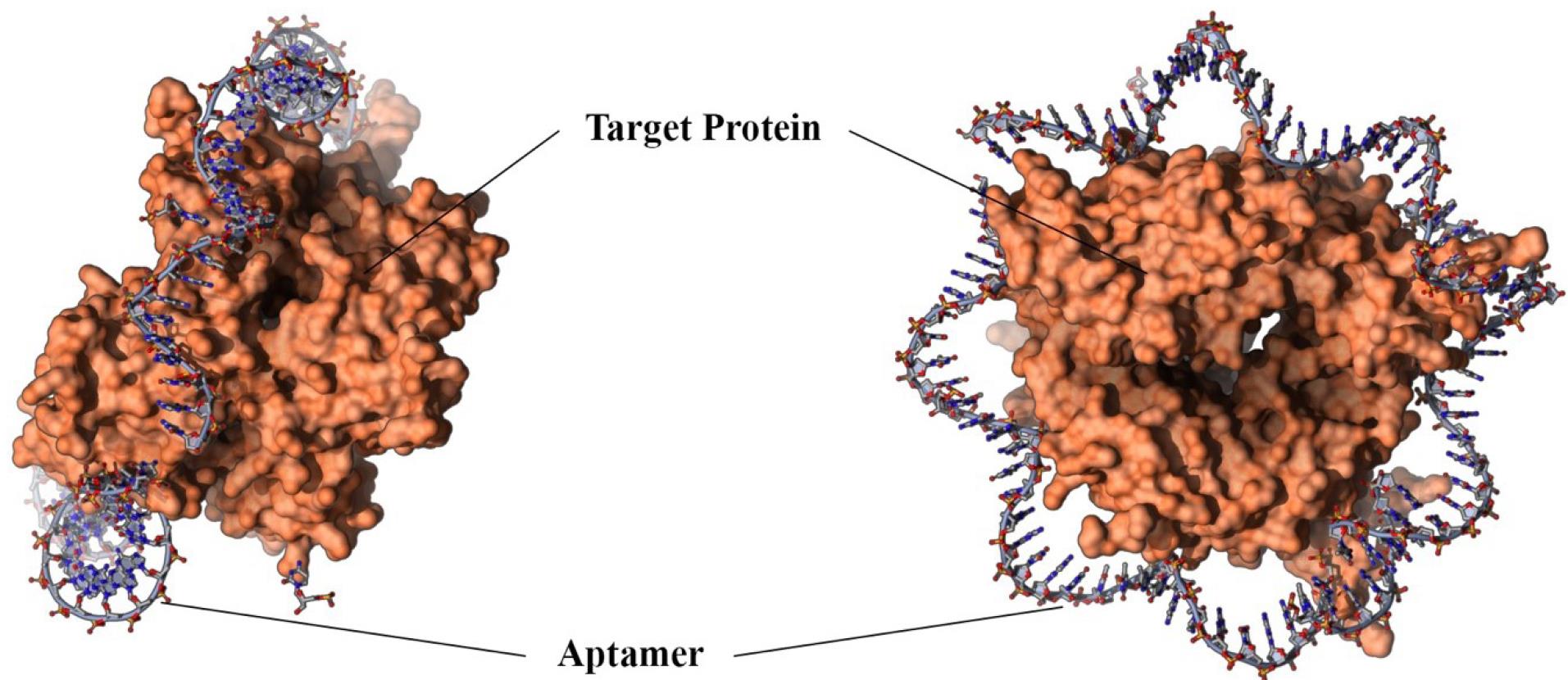
Figure 1.
Aptamer Structure
.
Aptamer Structure
General Applications of Aptamers
Aptamers have a variety of applications, but their common ones include the measurement of substances or the diagnosis and treatment of diseases. They have unique characteristics that make them better than other detector molecules, such as anticorps. Aptamers seem to be extremely smaller than enzymes and anticorps. In comparison with proteins, the phenomenon of non-specific absorption happens less often at nuclear acid levels.13 DNA aptamers are appropriate for the design of reusable and highly stable opto-sensors, whereas RNA aptamers provide a unit of measurement, and their main limitation is ribonuclease degradation, which is inhibited using diethyl pyrocarbonate-treated water.14
Immunogenicity Properties of Aptamers
Higher Stability Compared to Other Molecules
High temperatures cause proteins to lose their 3D structure, while oligonucleotides are thermally more stable. Aptamers change their structure without being degraded at temperatures above 60 °C and return to their original 3D structure when the temperature returns to room temperature, making aptamers a viable option for drug and vaccine immunization.15 The oligonucleotide aptamers were the most advantageous of all, and pharmaceutical researchers were persuaded to use aptamers as immunogens, in particular in the development of vaccines and other functional drugs.16 Aptamers have a denaturation-regeneration cycle and structure reversibility against various changes in physical conditions such as stress, temperature, pH, metal ions, and the like. While antibodies are unavailable in prepared forms, their denaturation is irreversible, and they are doomed to perish. Aptamers do not need to be stored at a low temperature and have a long shelf life.17 All these properties make aptamers good drugs since they can keep their immunogenic effects, such as activating defensive pathways, even in harsh environments that make other therapeutic molecules lose their functions.18 Figure 2 shows the Aptamer technology and its application at a glance.
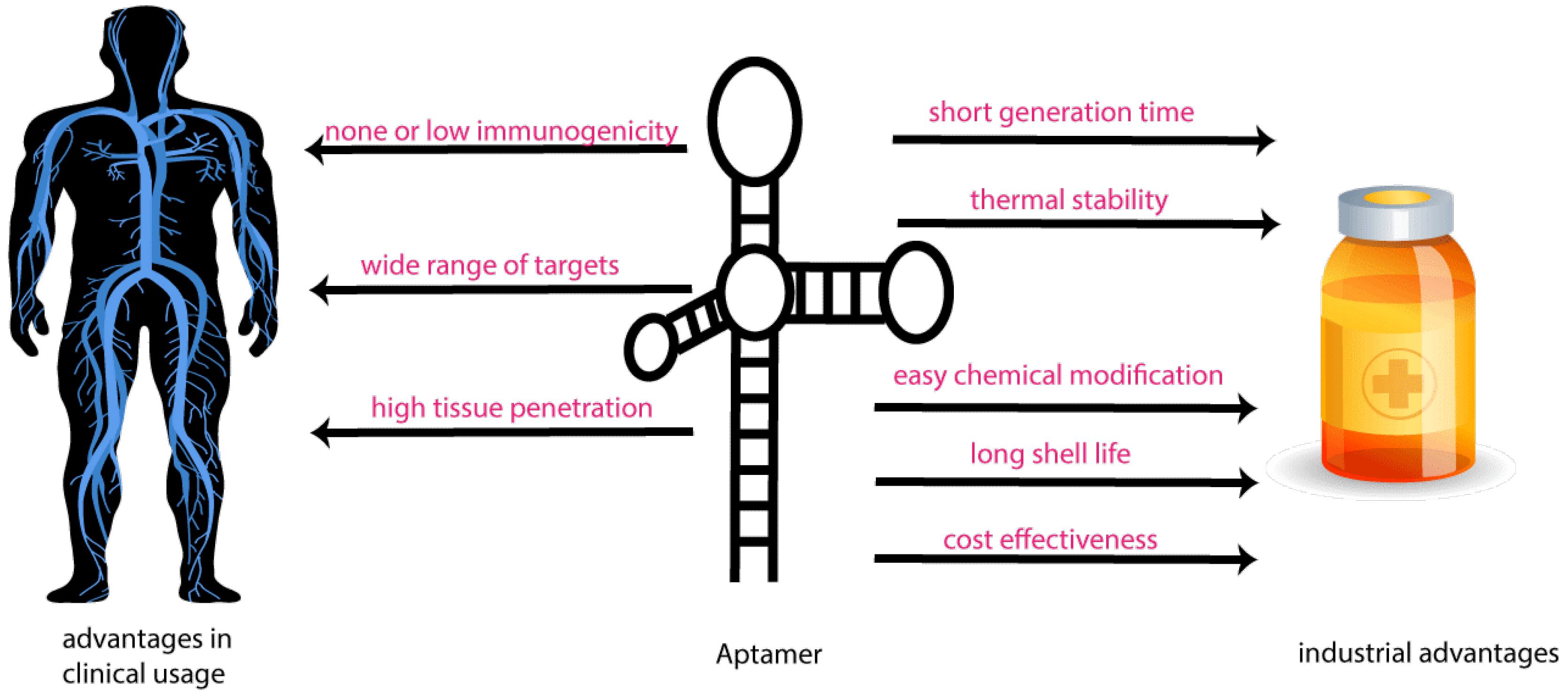
Figure 2.
Aptamer Technology and its Application
.
Aptamer Technology and its Application
Production of Aptamers
Monoclonal antibody detection and manufacturing is a time-consuming and expensive process that includes screening vast numbers of colonies. Furthermore, large-scale mammalian cell culture products are required for the financial success of antibodies. However, aptamers’ chemical synthesization is easy, cheap, and in vitro.19 While easily maintained, transported, and synthesized, aptamers, once chosen, have a high analytic affinity and specificity with low dissociation and reproducibility syntheses; in addition, their chemical process is highly precise. Further, the ability of aptamers to be easily altered by various chemical reactions enhances their stability and nuclease resistance. Because of these properties, aptamers have proven to be highly effective in the development of vaccines and immunization drugs.20 Another immunogenic feature of aptamers is their easy chemical manipulation, allowing for a broad spectrum of modifications and the addition of a wide range of functional and chemical groups. Moreover, it is effortless to regenerate and reproduce modified surfaces using aptamers.21
Synthesis of Aptamer Oligonucleotides for Immunogenicity
These aptamers are synthesized in the systematic evolution of ligands by the exponential enrichment (SELEX) cycle. This cycle is a practical approach to selecting high-quality aptamers from a library, including many random oligonucleotides, for both vaccine development and immunization. A library with 1013-1015 oligonucleotides is chemically generated in this cycle. These fragments each have an intermediate, random segment of approximately 60 nucleotides, which are utilized to select aptamers for medication and vaccine manufacture. They are selected with the best and most effective high immunogenic aptamer technique.13 Next, the library is used on a target (e.g., pure protein or cancer cells). In this way, aptamers with immunogenicity and unique properties get attached to the target.22 The connected parts are separated from the unconnected parts and then copied and given new targets as a new library. This cycle is repeated several times until the final selection, extension, and sequence are made of the most specific components with high immunogenicity. Over the years, this technique has been modified and improved at various stages to reduce the number of cycles and its duration in its new types 23. The procedure is illustrated in Figure 3.
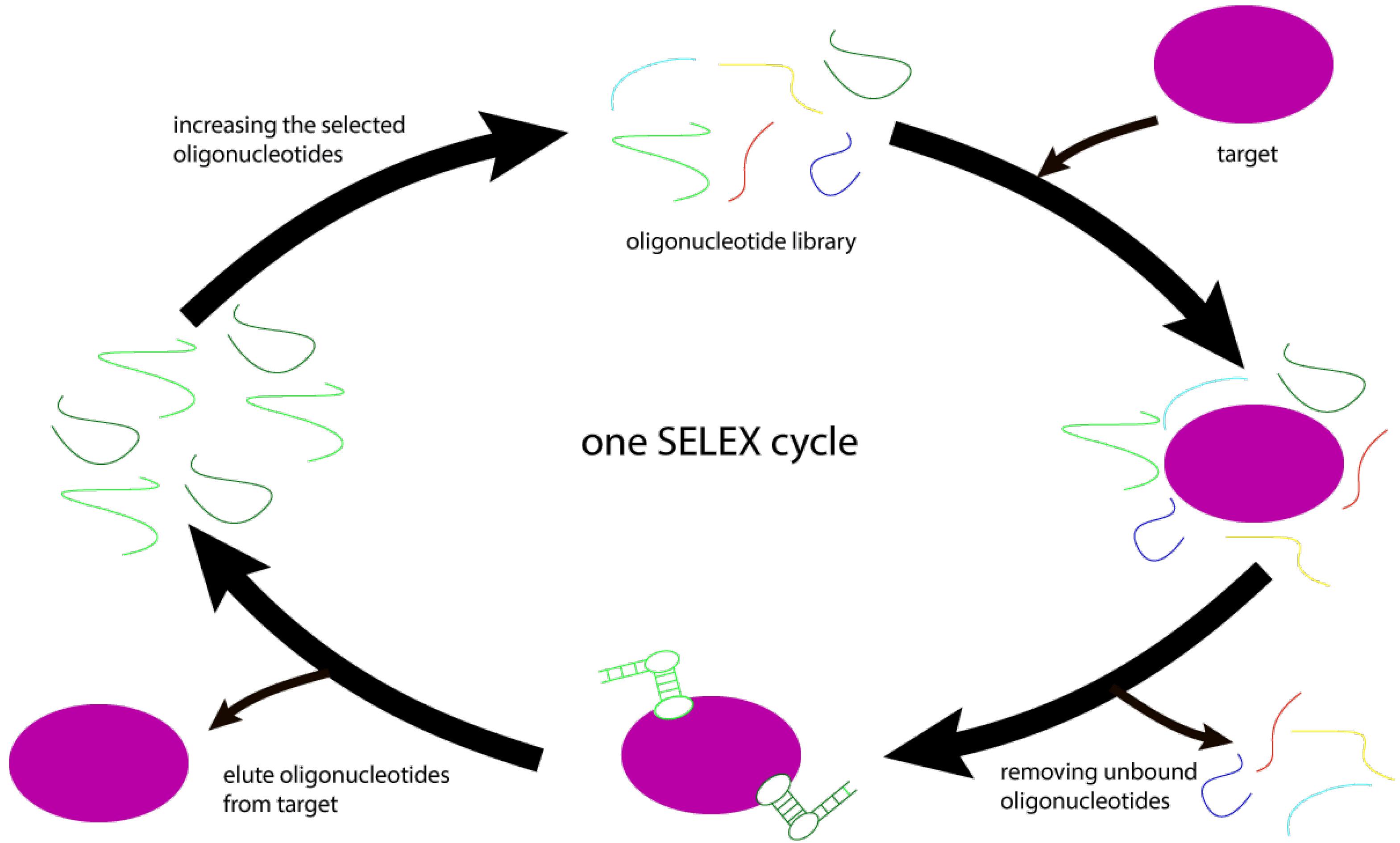
Figure 3.
Synthesis of High Immunogenicity Aptamer Oligonucleotides
.
Synthesis of High Immunogenicity Aptamer Oligonucleotides
Immunogenic Application of Aptamers
Due to their oligonucleotide properties, such as their high specificity and purpose-related affinity, low cost of production, and easy binding to reporter molecules, they can replace antibodies in numerous fields, particularly in the pharmaceutical and vaccine industries. These programs involve identifying and processing diseases, biosensors, and drug carriers.23 For instance, in a program, two aptamers were developed that had anti-CD28 effects. CD28 is one of the main costimulatory receptors responsible for the proper activation of T lymphocytes. The program proved that these aptamers can be easily engineered, depending on the needs of the immunological approach, to function as either antagonistic (blocking) or agonistic (activating) by means of a simple dimerization.24 Finally, it showed that agonistic anti-CD28 aptamers boost the antitumor immune response elicited by idiotypic vaccination in a murine lymphoma model, increasing the overall survival to a considerable extent.25
Aptamers in Pharmaceutical Biotechnology and Pharmacy
The SELEX technique is used to select aptamers from a huge number of oligonucleotide sequences. Aptamers have several benefits over antibodies, which are provided as follows:
Unlike antibodies, aptamers are produced in vitro using chemical procedures. Unlike antibodies, they detect a broad spectrum of target molecules, such as proteins, peptides, toxins, amino acids, drugs, metal ions, and even complete cells.26 In addition, chemical modification in aptamers is easier to do compared to antibodies. Further, unlike antibodies, aptamers’ denaturation, which happens due to an increase in temperature, is reversible. Furthermore, aptamers can be more densely immobilized at the bioreceptor level in biosensors, which increases their sensitivity in identifying and measuring the target molecule. Moreover, many aptamers change conformations by binding to the target molecule, a feature that aids in the signal generation process in some biosensor systems. Aptamer applications are depicted in Figure 4.
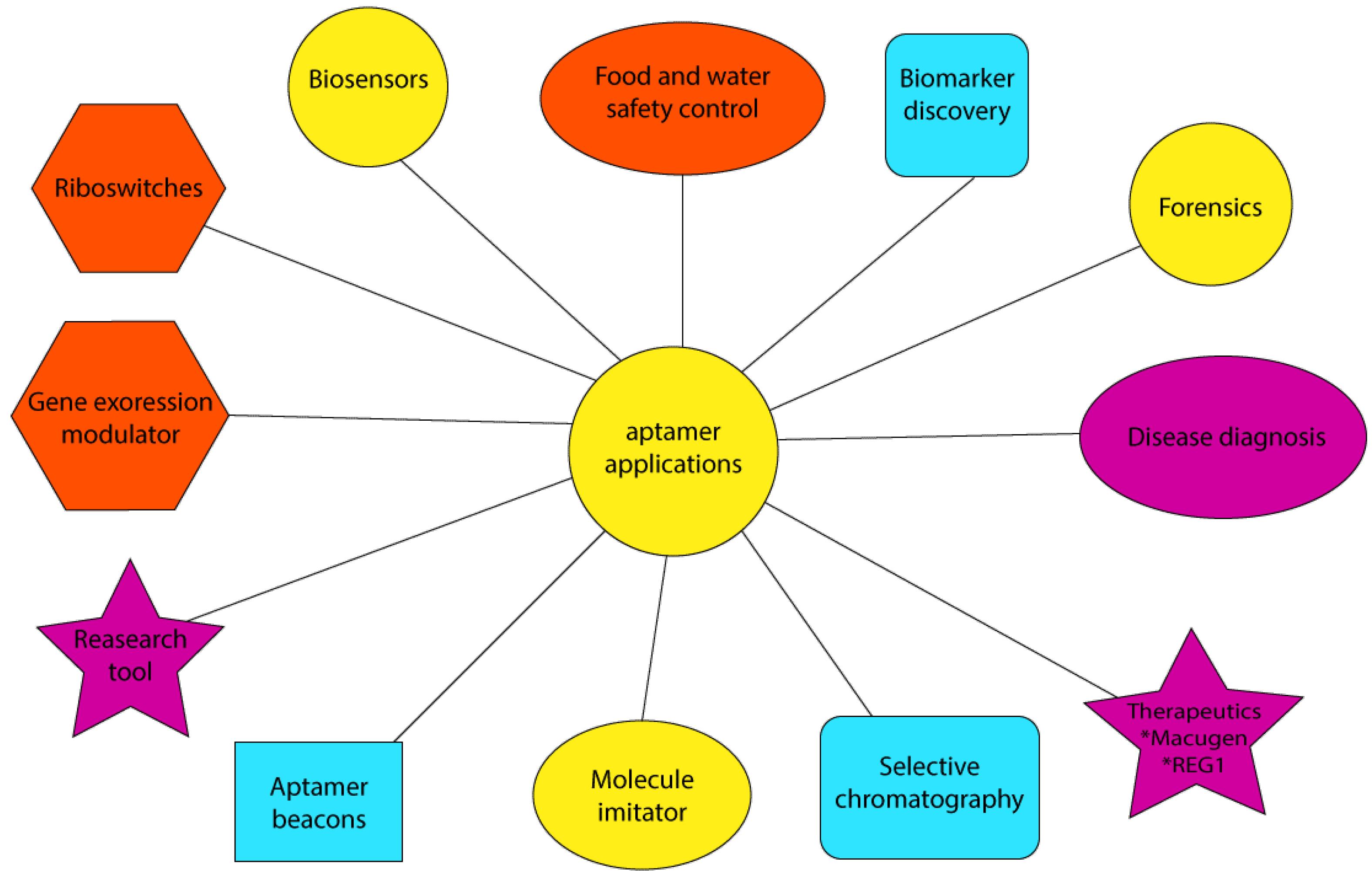
Figure 4.
Application of Aptamers in Pharmaceutical Biotechnology and Pharmacy
.
Application of Aptamers in Pharmaceutical Biotechnology and Pharmacy
Two types of target molecules are recognized by aptamers, including small and large (e.g., proteins) molecules. Aptamer is folded into a 3D structure in such a way that slits are created on its surface for the connection of small target molecules. As regards large molecules, the aptamer is folded in such a way that it takes place in the gaps and surface grooves of the large molecule.27 In the field of biotechnology, considerable attention is dedicated to aptamers because of the benefits of antibodies. These molecules may be employed as drugs to design biosensors and supply them to inhibit a particular protein, enzyme, or the like.28
Acid-nucleic Aptamers: New Immunogenic Tools in the Process of Diagnosis and Treatment
Aptamers bind to specific targets, including proteins, peptides, and carbohydrates. Aptamers recognize targets and bind to them with a 3D structure, while antibodies recognize the linear epitopes of the targets. Considering that they are chemically synthesized, the process can simply be scaled up, not to mention that the batch-to-batch variation of the process is low, and the product is highly stable.29 Additionally, the production of aptamers is fast and cost-effective. Aptamers are highly safe biomaterials with no reported in vivo immunogenicity. This review article addresses the production method of nucleic acid aptamers, their advantages and disadvantages in comparison to antibodies, and their prospective applications in medicine (Figure 5). Aptamers’ limitations and strategies to overcome them also underwent investigation.30
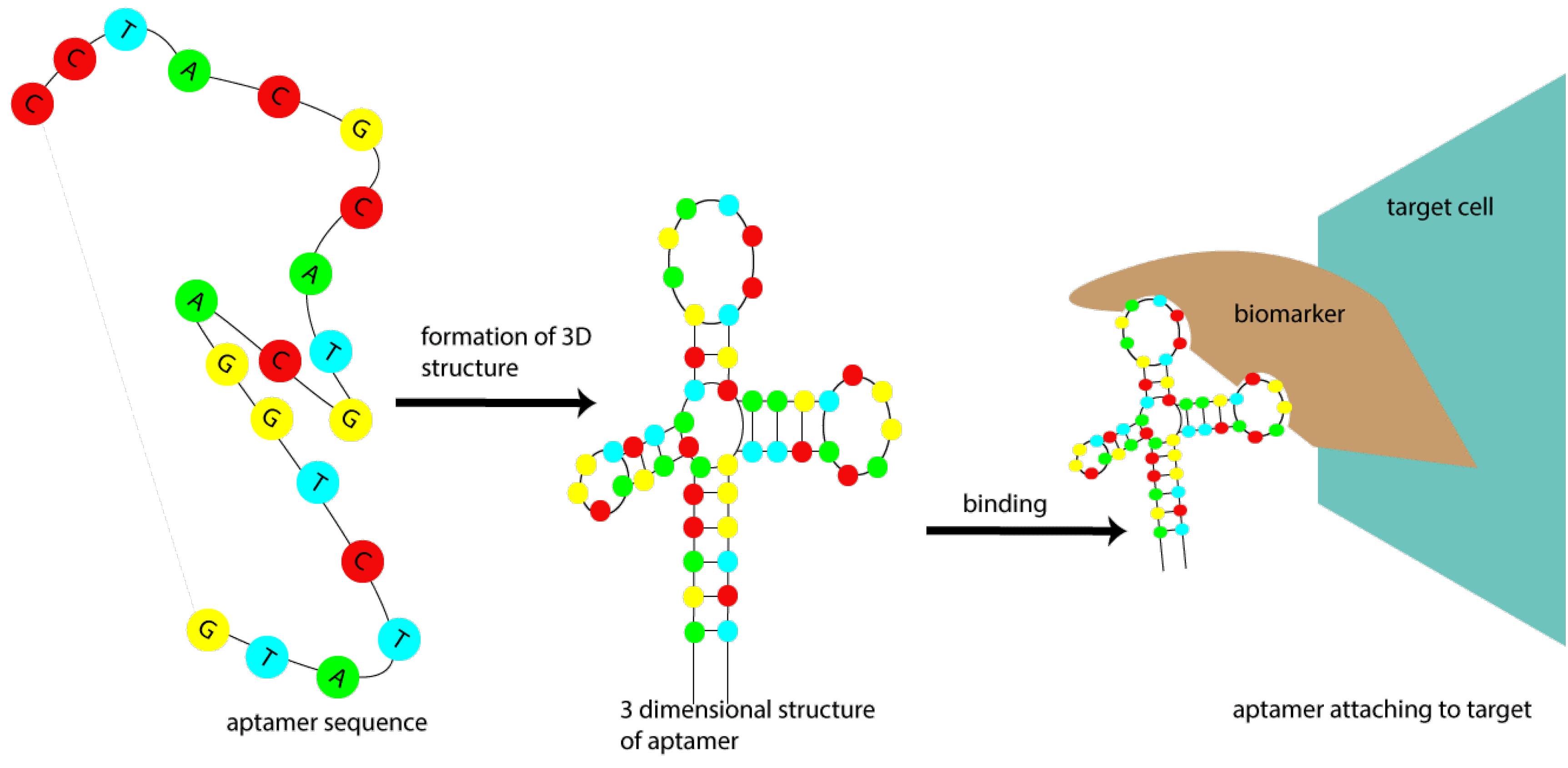
Figure 5.
Schematic Diagram of Aptamer Function
.
Schematic Diagram of Aptamer Function
Isolation and Selection of High-Affinity Aptamer DNA Against Vascular Endothelial Growth Factor (Angiogenic Factor) for High Immunogenicity
VEGF is a significant growth factor in the cell division and differentiation of tissue, which is needed for the synthesis and development of new blood vessels that are meant for the tissue transportation of oxygen and nutrients.31
As a result, this growth factor can be employed to stimulate angiogenesis in tissues using in vivo methods to treat damaged body tissues and in vitro methods for tissue engineering in medicine. On the other hand, in some situations, the body produces this component in an excessive and unregulated manner, which occurs in disorders and diseases such as age-related macular degeneration (AMD), diabetic macular edema, and the appearance of malignant tumors.32 VEGF contains several different isoforms within the body. VEGF165 is the most abundant and noticeable isoform in the body, which plays a vital role in the process of tissue angiogenesis.33 VEGF165 is also a helpful biomarker for cancer tumor diagnostics due to its increased cancer cell secretion. There is, therefore, a considerable necessity for cleansing and isolating this protein factor to use as a growth factor necessary for angiogenesis in the tissues and inhibiting its secretion or production to treat disorders caused by its unregulated secretion. Antibodies are one of the most important ligands utilized for protein isolation in chromatography while being used as medications.34 However, there are some limitations to using antibodies, including immunogenicity, high biodegradation and instability, and high production costs. Today, novel nucleic acid ligands known as aptamers are utilized to solve this issue. The aptamer is a short single-stranded oligonucleotide sequence of RNA or DNA with a specific 3D form and the ability to attach specifically to the target protein with high affinity. In contrast to common anticorps, aptamers are not developed in biological systems but may be produced easily in the laboratory.35 The perfect example of the mentioned aptamers would be Pegaptanib, which exerts several physiological and pathological functions, such as an increase in vascular permeability causing fluid leakage in wet AMD.36 Pegaptanib selectively interacts with the heparin-binding site of the extracellular VEGF165 isoform and prevents VEGF165 from stimulating its receptor on the surface of the endothelial cell, thus blocking the initiation of the intracellular cascade and consequently inhibiting vascular permeability and retina neovascularization.37
Selection of Aptamers by Systematic Evolution of Ligands by Exponential Enrichment Technology for Increased Drug Immunogenicity
The first step in the design of opt censors is the identification of a specific aptamer for the target molecule by an adaptive selection process called SELEX.38 In the 1980s, human immunodeficiency investigations were performed on the concept of protein binding to DNA. Studies have revealed that such viruses encode a variety of short RNAs that bind to cellular or viral proteins with high specificity and affinity. The SELEX process at the Goldo Zhostak Laboratory was reported in subsequent investigations in 1990. Aptamers are selected randomly from a synthesized oligonucleotide library during the SELEX procedure. SELEX is a hybrid method of chemistry that selects a single string from a library of 1015 to 1016 oligonucleotides with a length of 40 to 100 bp.39 Each strand is comprised of two fixed segments for amplification by polymerase chain reaction (PCR) on both sides of the oligonucleotides and one segment between them in a random variable sequence. In each SELEX procedure, a random DNA oligonucleotide library is developed, which is used directly for selecting DNA aptamers.40 This library must, first, be transformed into an RNA library to choose RNA aptamers. SELEX is defined by the repetition of five successive steps, including connection, separation, rinse, amplification, and conditioning. The library and target molecule will be incubated to connect in the first round of the SELEX procedure. Unbound oligonucleotides are removed by multiple vigorous wash steps from the connected complex.41 The oligonucleotides are washed before being amplified by RT-PCR or PCR. In this situation, a new enriched reservoir of selected oligonucleotides is produced by the PCR transcription of the suitable ssDNA from the SELEX results.42 This selected oligonucleotide resource is then used in the subsequent round of selection. To select specific aptamers for high-affinity target molecules, 6-20 rounds of the SELEX procedure are often necessary.43 Figure 6 sums up the SELEX procedure.
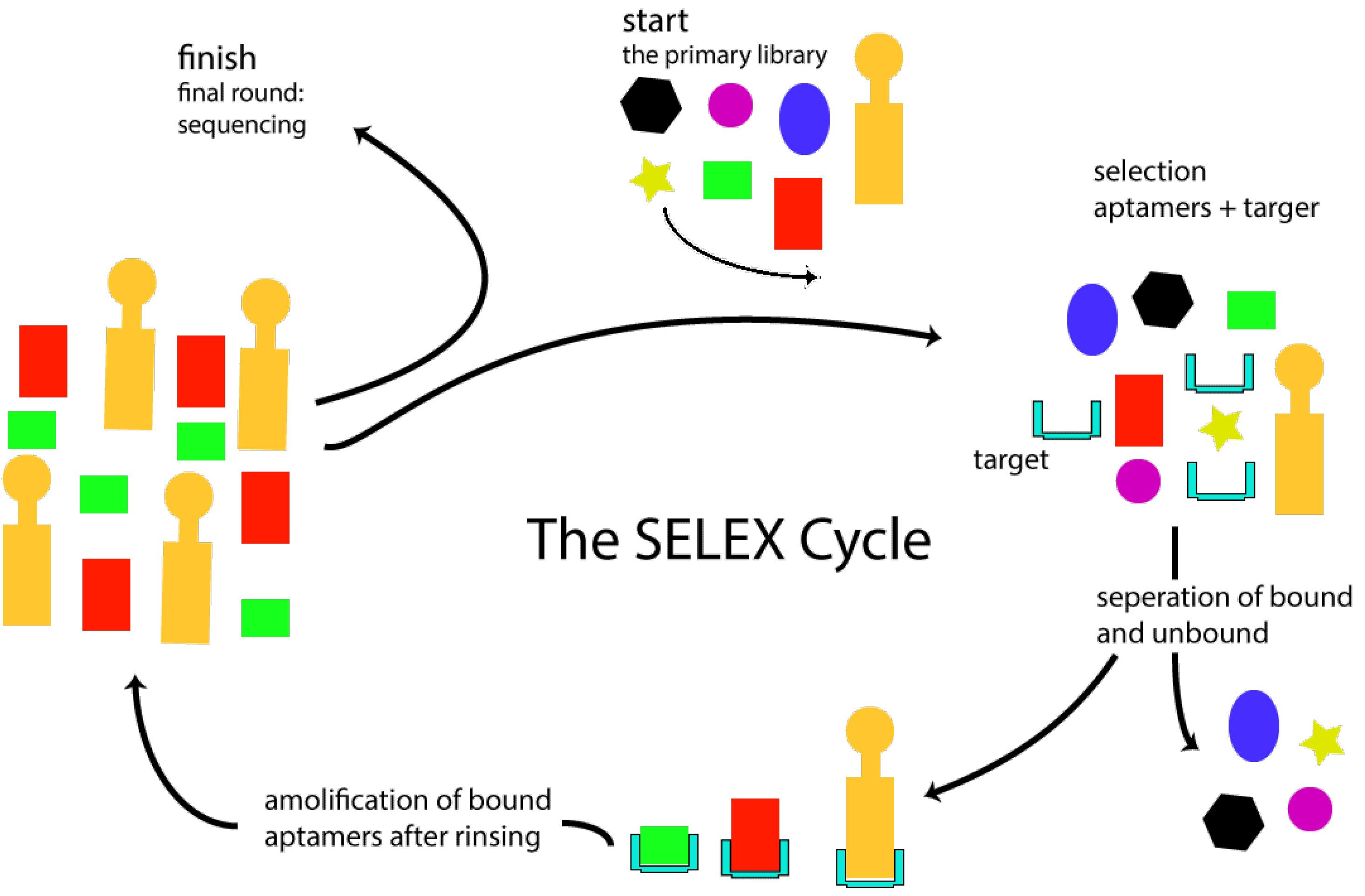
Figure 6.
Selection of Aptamers With High Safety and Special Properties by SELEX Technology. Note. SELEX: Systematic evolution of ligands by exponential enrichment
.
Selection of Aptamers With High Safety and Special Properties by SELEX Technology. Note. SELEX: Systematic evolution of ligands by exponential enrichment
Despite the tremendous growth in the usage of aptamers for decomposition work as molecular detection elements, their interaction with target molecules necessitates the employment of a digital decomposition system. In reality, for the optimal usage of aptamer as a parser probe, it is crucial to establish a quantifiable signal from aptamer-target molecule interaction by simple approaches 44. To date, multiple methods have been developed for measuring distinct molecules’ properties, such as light transmission, mass-sensitive mass, or electrochemical measurements.45 Many biosensors, however, are generated using electrochemical techniques. A biosensor is an analytical instrument in electrochemistry that transforms biological responses into electric output.46
DNA Aptamer Immunogenicity
Many 3D compounds are formed by nucleic acid sequences such as DNA and RNA. Some of these structural variants can attach to proteins and tiny molecules. Oligonucleotides with adequate binding properties can be screened using a practical selection approach.47 Many pieces of research on the potential laboratory applications of aptamers have been published lately. According to these findings, aptamers play a crucial role in immunogenicity. Aptamers may be viable alternatives to immunological reagents and immuno-oligonucleotide compounds in some methods.48
Effect of Aptamer on Immunoglobulin (as an Immunogenic Effect)
The interaction between aptamer RNA and human immunoglobulin G (IgG) has been identified by Japanese researchers. Aptamer Apt8-2 is a molecule that is 23 nucleotides long. This aptamer also contains two fluoropyrimidines that bind to the FC segment in human IgG with considerable affinity.49 However, this aptamer is unable to bind to the animal IgG. Apt8-2 competes with a protein for binding to FC in IgG. Apt8-2 competes with protein A, but not with the Fcγ receptor, for IgG binding. Nuclear magnetic resonance chemical-shift analyses localize the aptamer-binding sites on the Fc subdomain, which partially overlaps the protein A binding site rather than the Fcγ receptor binding site. The tertiary structures of the predicted recognition sites on the Fc domain differ significantly between human IgG and other species of IgGs; this, in part, accounts for the high specificity of the selected aptamer. Apt8-2 can, therefore, be used as a protein A alternative for the affinity purification of human IgG and therapeutic antibodies. Apt8-2 interaction with FC is detected only in the presence of Mg+2 and Ca+2 (The Apt8-2 response to FC is only noticed when Mg+2 and Ca+2 are present). The aptamer bound to the IgG is released by removing these ions with ethylenediaminetetraacetic acid (EDTA). Acid buffers are also utilized to release IgG from protein A. As a result, acidic conditions can be deeply destructive to IgG. For this reason, it is recommended that Apt8-2 be utilized to release IgG because EDTA is used to extract IgG from Apt8-2.50
Aptamer Reaction With Receptor Cells in the Immunogenic Process
Aptamer was previously employed to modulate a wide range of cellular activities in immune responses.51 Aptamer has the potential to block various cellular receptors and prevent their interactions with ligands. Various investigations have also revealed that aptamers can behave as stimulators of isolated reactions. Xiang et al combined different aptamers to minimize immunological reactions.52 In other trials, they could use aptamers that bind to proteins such as CD-28, which is involved in lymphocyte T activation. Many aptamers are capable of blocking the B7 connection to CD 28. Some anti-CD 28 aptamers can also function as agonists, triggering the anti-tumor cellular immune response.53 In addition, the anti-CD28 aptamers, which have been designed, show two different functions depending on which is needed, one being antagonistic (blocking) or agonistic (activating), which is related to dimerization. It has been demonstrated that the agonistic function can increase the immunogenicity of vaccination in a murine lymphoma model, resulting in higher survival rates.54
Combination of Aptamer With Soluble Proteins
C5 is a part of the innate immune system that aids in the response against infectious pathogens. C5 proteolytic components are found in highly active inflammatory proteins. In C5, the proteolytic components are a portion of the inflammatory proteins that are highly active.55 C5a is a major cause of systemic inflammation in a variety of diseases, including sepsis and autoimmune disorders. Multiple types of aptamers have been synthesized to restrain C5 and its proteolytic components in this regard. The C5 protein binds various RNA molecules with dissociation constants comparable to those of M1 RNA, the RNA component of RNase P, one of the major ones being the W2 sequence. The W2 sequence has independent mg2+ interactions with C5, though the affinity between them increases with an elevated level of NH4+ in the environment, suggesting the existence of hydrophobic bonds between them.56 The core RNA motif essential for interaction with the C5 protein was identified as a stem–loop structure, comprising a 5 bp stem and a 20 nt loop. Such aptamers as W2 can protect cells from proteolytic destruction by binding to C5 components. Nuclease-resistant aptamers (RNA/DNA NOX-D20) are synthesized from aberrant oligonucleotides. In humans and mice, RNA/DNA NOX-D20 can suppress C5 activity.57
Immunogenic and Therapeutic Applications of Aptamers
Application of Aptamer in the Field of Treatment
This section discusses aptamers that have entered the realm of treatment and are in various stages of clinical trials (Table 1). The first of these aptamers is Macugen, which has been invented by Pfizer and Eyetech and is currently commercially available for the treatment of AMD 58. It is an aptamer conjugated with polyethylene glycol, a single-stranded nucleic acid specifically for VEGF-165, which plays a key role in angiogenesis and permeability.59
Table 1.
Clinical Level Aptamers
|
Aptamer Name
|
Target
|
Indication
|
Clinical Phase
|
| Macugen |
VEGF-165 |
Angiogenesis and permeability |
Clinically approved |
| REG1 |
Coagulation factor IXa |
Applied as an anticoagulant |
Phase III |
| AS1411 |
Nucleolin |
Acute myeloid leukemia |
Phase II |
| ARC1779 |
von Willebrand factor |
Carotid artery disease |
Phase 1 |
| NU172 |
thrombin |
Applied as an anticoagulant |
Phase II |
| NOX-E36 |
CCL2 (C-C Chemokine Ligand 2)/ MCP-1 (monocyte chemoattractant protein 1) |
Type 2 diabetes mellitus |
Phase II completed |
| NOX-A12 |
CXCL12 (C-X-C Chemokine Ligand 12) |
Metastatic colorectal cancer |
Phase II |
| NOX-H94 |
Hepcidin |
Anemia of chronic diseases |
Phase II |
| E10030 |
PDGF |
AMD Von Hippel-Lindau |
Phase III |
| ARC19499 |
Tissue factor pathway inhibitor |
Hemophilia |
Phase 1 completed |
Note. VEGF: Vascular endothelial growth factor; PDGF: Platelet-derived growth factor; AMD: Age-related macular degeneration.
REG1
It was invented as a novel aptameric anticoagulant drug. This drug (Figure 7) is an aptamer in phase two of clinical trials. It is made up of RB006 (coagulation factor 9-specific aptamer) and RB007 (aptamer RB006 oligonucleotide antidote).60 RB006 is a 2-ribopurine/2-fluoropyrimidine aptamer conjugated to 40 kDa polyethylene glycol to protect it from nuclease degradation.61
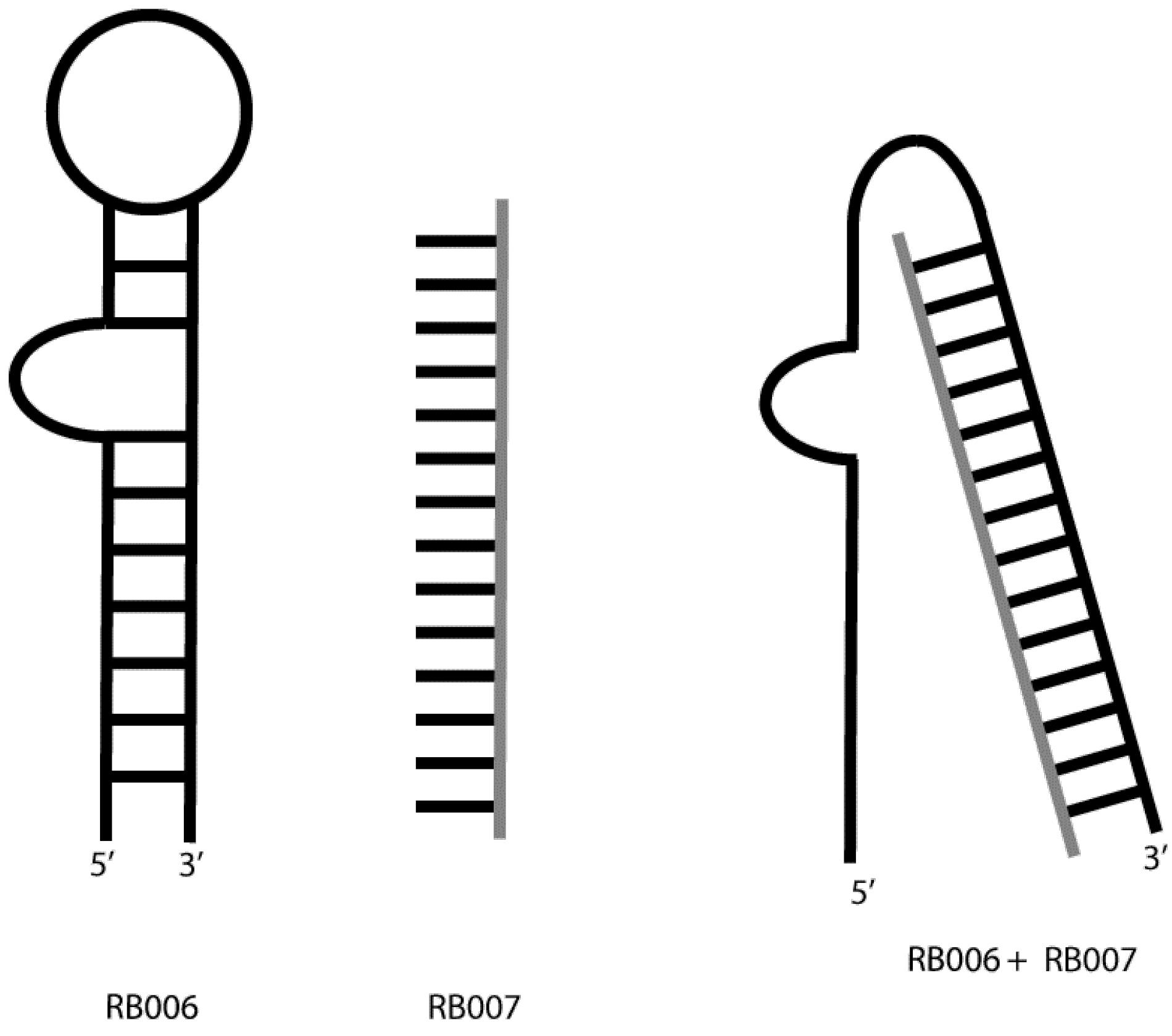
Figure 7.
REG1 Aptamer Structure
.
REG1 Aptamer Structure
AS1411
A detailed representation is displayed in Figure 8. It is a nucleolin-specific aptamer that is used to treat acute myeloid leukemia.62
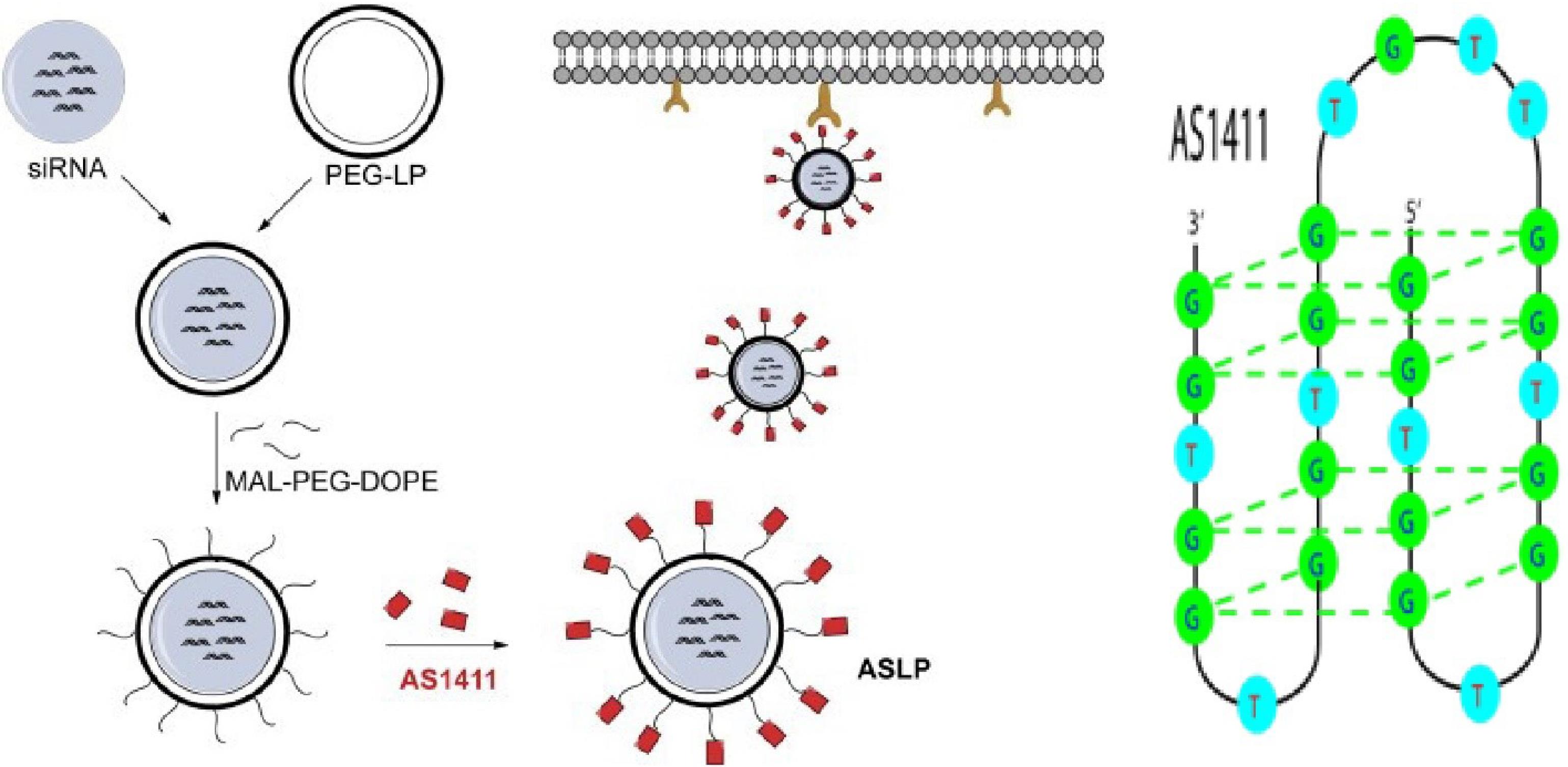
Figure 8.
AS1411 Aptamer Structure
.
AS1411 Aptamer Structure
ARC1779
ARC1779 (Figure 9) is a proprietary aptamer of the von Willebrand factor. This aptamer is prescribed to patients suffering from carotid artery disease.
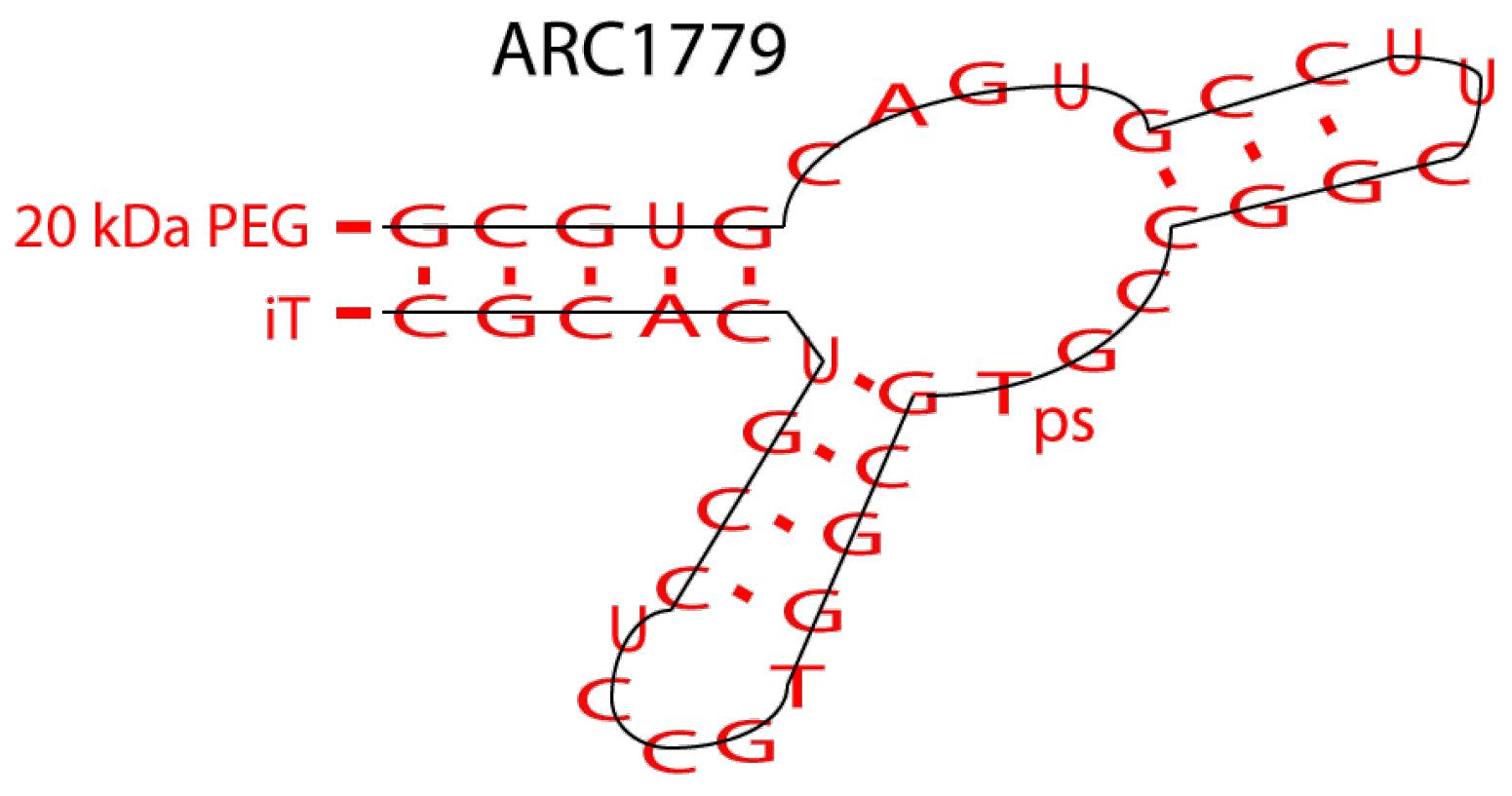
Figure 9.
ARC1779 Aptamer Structure
.
ARC1779 Aptamer Structure
NU172
It is a particular aptamer of thrombin, applied in various clinical stages as an anticoagulant (Figure 10).63
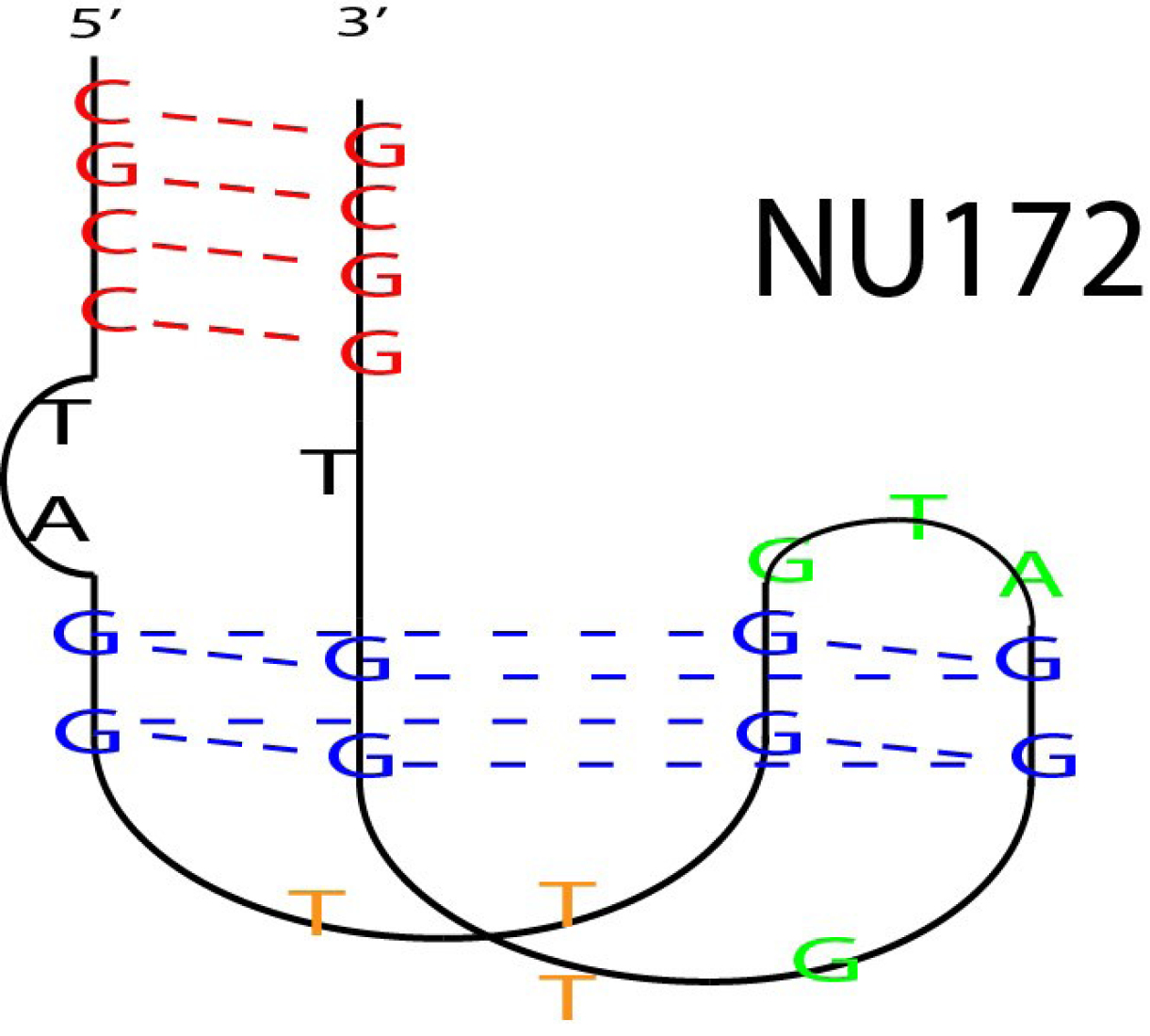
Figure 10.
NU172 Aptamer Structure
.
NU172 Aptamer Structure
Diagnostic Programs of Aptamers for Drug Safety
Aptamers can be applied to biomass through chemical modifications, either with luminescence or by binding to different nanoparticles (NPs). In one study, for example, DNA aptamer against Bacillus anthracis spores was produced in the form of a sandwich (an electrochemiluminescence magnetic sphere aptamer) to detect anthrax spores.64,65 Another diagnostic application is the identification of cancer cell biomarkers through the production of aptamers adverse to these markers, which are used to detect and image malignant tissue.59
Conclusion
RNA self-assembling of diverse therapeutic oligonucleotide NPs has so far achieved tremendous progress in nanotechnology. These NPs are generally composed of RNA molecules (aptamers) and have great potential for therapeutic applications because of their features. Some of these characteristics are appropriate size, multiple capacity, minimal stimulation of the immune response of molecular production, intrinsic RNA, chemical nature of NPs through chemical synthesis with infinite accuracy and reproducibility, high-grade RNA purification, rapid return of drug activity, and strength.
In this paper, some methods were presented for the production and selection of aptamers, along with a list of the advantages and disadvantages of these agents and their immunogenicity.52 Aptamers have advantages over antibodies that have created a new perspective for their use in diagnostic and therapeutic methods. Antibodies produced against the target molecule bind to the target protein only under physiological conditions (a narrow range of temperature, acidity, and ionic strength), whereas aptamers are not restricted in this way.53
Aptamers have advantages over antibodies that have created a new perspective for their use in diagnostic and therapeutic methods. In principle, aptamer-based NPs and their varied immunogenicity will have different applications, and it is reasonable to conclude that nanotechnology will play a crucial role in expediting translation and development in the years ahead.54
Authors’ Contribution
Conceptualization: Tooba Gholikhani.
Data curation: Tooba Gholikhani.
Funding acquisition: Tooba Gholikhani.
Investigation: Shalen Kumar.
Project administration: Tooba Gholikhani.
Resources: Amirhossein Olfati.
Software: Shalen Kumar.
Supervision: Tooba Gholikhani.
Validation: Arin Ghazizadeh, Raha Amazadeh, Mahyar Ziyamolki.
Visualization: Mahyar Ziyamolki.
Writing–original draft: Arin Ghazizadeh, Raha Amazadeh.
Writing–review & editing: Shalen Kumar.
Ethical Approval
The study was arroved by Ethics Committees of Vice-Chancellor in Research Affairs - Tabriz University of Medical Sciences (Code: IR.TBZMED.VCR.REC.1399.420).
Funding
Self fund.
References
- Chauveau F, Pestourie C, Ducongé F, Tavitian B. Aptamer selection by Darwinian evolution. In: Boisseau P, Houdy P, Lahmani M, eds. Nanoscience: Nanobiotechnology and Nanobiology. Berlin, Heidelberg: Springer; 2009. p. 223-49. 10.1007/978-3-540-88633-4_6.
- Gholikhani T, Kumar S, Valizadeh H, Mahdinloo S, Adibkia K, Zakeri-Milani P. Advances in aptamers-based applications in breast cancer: drug delivery, therapeutics, and diagnostics. Int J Mol Sci 2022; 23(22):14475. doi: 10.3390/ijms232214475 [Crossref] [ Google Scholar]
- Gholikhani T, Jimenez Brito B, Livingston F, Kumar S. The potential use of aptamers in the process of drug development. Pharm Sci 2021; 27(4):503-10. [ Google Scholar]
- Charkhpour M, Delazar A, Mohammadi H, Gholikhani T, Parvizpur A. Evaluation of the effects of Artemisia austriaca on morphine withdrawal syndrome in rats. Pharm Sci 2014; 20(1):1-5. [ Google Scholar]
- Zakeri-Milani P, Najafi-Hajivar S, Sarfraz M, Nokhodchi A, Mohammadi H, Montazersaheb S. Cytotoxicity and immunogenicity evaluation of synthetic cell-penetrating peptides for methotrexate delivery. Iran J Pharm Res 2021; 20(3):506-15. doi: 10.22037/ijpr.2021.114429.14842 [Crossref] [ Google Scholar]
- Memar MY, Ahangarzadeh Rezaee M, Barzegar-Jalali M, Gholikhani T, Adibkia K. The antibacterial effect of ciprofloxacin loaded calcium carbonate (CaCO3) nanoparticles against the common bacterial agents of osteomyelitis. Curr Microbiol 2023; 80(5):173. doi: 10.1007/s00284-023-03234-y [Crossref] [ Google Scholar]
- Khajeei A, Masoomzadeh S, Gholikhani T, Javadzadeh Y. The effect of PEGylation on drugs’ pharmacokinetic parameters; from absorption to excretion. Curr Drug Deliv. 2023. 10.2174/1567201820666230621124953.
- Zhu Q, Liu G, Kai M. DNA aptamers in the diagnosis and treatment of human diseases. Molecules 2015; 20(12):20979-97. doi: 10.3390/molecules201219739 [Crossref] [ Google Scholar]
- Kimoto M, Yamashige R, Matsunaga K, Yokoyama S, Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat Biotechnol 2013; 31(5):453-7. doi: 10.1038/nbt.2556 [Crossref] [ Google Scholar]
- Chen Z, Wu Q, Chen J, Ni X, Dai J. A DNA aptamer based method for detection of SARS-CoV-2 nucleocapsid protein. Virol Sin 2020; 35(3):351-4. doi: 10.1007/s12250-020-00236-z [Crossref] [ Google Scholar]
- Lakhin AV, Tarantul VZ, Gening LV. Aptamers: problems, solutions and prospects. Acta Naturae 2013; 5(4):34-43. [ Google Scholar]
- Mou Q, Xue X, Ma Y, Banik M, Garcia V, Guo W. Efficient delivery of a DNA aptamer-based biosensor into plant cells for glucose sensing through thiol-mediated uptake. Sci Adv 2022; 8(26):eabo0902. doi: 10.1126/sciadv.abo0902 [Crossref] [ Google Scholar]
- Xu G, Wang C, Yu H, Li Y, Zhao Q, Zhou X. Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer. Nucleic Acids Res 2023; 51(14):7666-74. doi: 10.1093/nar/gkad541 [Crossref] [ Google Scholar]
- Yan Y, Chang D, Xu Y, Chang Y, Zhang Q, Yuan Q. Engineering a ligase binding DNA aptamer into a templating DNA scaffold to guide the selective synthesis of circular DNAzymes and DNA aptamers. J Am Chem Soc 2023; 145(4):2630-7. doi: 10.1021/jacs.2c12666 [Crossref] [ Google Scholar]
- Villa A, Brunialti E, Dellavedova J, Meda C, Rebecchi M, Conti M. DNA aptamers masking angiotensin converting enzyme 2 as an innovative way to treat SARS-CoV-2 pandemic. Pharmacol Res 2022; 175:105982. doi: 10.1016/j.phrs.2021.105982 [Crossref] [ Google Scholar]
- Silwal AP, Thennakoon SKS, Arya SP, Postema RM, Jahan R, Phuoc CMT. DNA aptamers inhibit SARS-CoV-2 spike-protein binding to hACE2 by an RBD- independent or dependent approach. Theranostics 2022; 12(12):5522-36. doi: 10.7150/thno.74428 [Crossref] [ Google Scholar]
- Li R, Wu X, Li J, Lu X, Zhao RC, Liu J. A covalently conjugated branched DNA aptamer cluster-based nanoplatform for efficiently targeted drug delivery. Nanoscale 2022; 14(26):9369-78. doi: 10.1039/d2nr01252a [Crossref] [ Google Scholar]
- Saito T, Shimizu Y, Tsukakoshi K, Abe K, Lee J, Ueno K. Development of a DNA aptamer that binds to the complementarity-determining region of therapeutic monoclonal antibody and affinity improvement induced by pH-change for sensitive detection. Biosens Bioelectron 2022; 203:114027. doi: 10.1016/j.bios.2022.114027 [Crossref] [ Google Scholar]
- Liu Z, Xue J, Chen L, Ma L, Yang H, Zhang Y. A signal-off aptamer sensor based on competition with complementary DNA and click polymerization for electrochemical detection of AFB1. Microchem J 2022; 181:107775. doi: 10.1016/j.microc.2022.107775 [Crossref] [ Google Scholar]
- Fujita K, Honda T, Tsukakoshi K, Ohno H, Ikebukuro K. The state of water molecules induces changes in the topologies and interactions of G-quadruplex DNA aptamers in hydrated ionic liquid. J Mol Liq 2022; 366:120175. doi: 10.1016/j.molliq.2022.120175 [Crossref] [ Google Scholar]
- Vijayakumar S, Nasr SH, Davis JE, Wang E, Zuidema JM, Lu YS. Anti-KIT DNA aptamer-conjugated porous silicon nanoparticles for the targeted detection of gastrointestinal stromal tumors. Nanoscale 2022; 14(47):17700-13. doi: 10.1039/d2nr03905b [Crossref] [ Google Scholar]
- Du Y, Wang Q, Shi L, Li T. G-quadruplex-proximized aptamers (G4PA) efficiently targeting cell-surface transferrin receptors for targeted cargo delivery. Nano Lett 2022; 22(15):6328-33. doi: 10.1021/acs.nanolett.2c02064 [Crossref] [ Google Scholar]
- Lin Y, Xu Y, Xing Y, Liu N, Chen X. Photoreversible DNA nanoswitch-based eluent-free strategy for the direct and effective isolation of highly-active thrombin from whole blood. Int J Biol Macromol 2023; 239:124359. doi: 10.1016/j.ijbiomac.2023.124359 [Crossref] [ Google Scholar]
- Guo W, Gao H, Li H, Ge S, Zhang F, Wang L. Self-assembly of a multifunction DNA tetrahedron for effective delivery of aptamer PL1 and Pcsk9 siRNA potentiate immune checkpoint therapy for colorectal cancer. ACS Appl Mater Interfaces 2022; 14(28):31634-44. doi: 10.1021/acsami.2c06001 [Crossref] [ Google Scholar]
- Ozer I, Pitoc GA, Layzer JM, Moreno A, Olson LB, Layzer KD. PEG-like brush polymer conjugate of RNA aptamer that shows reversible anticoagulant activity and minimal immune response. Adv Mater 2022; 34(10):e2107852. doi: 10.1002/adma.202107852 [Crossref] [ Google Scholar]
- Yaghoobi E, Zavvar T, Ramezani M, Alibolandi M, Rahimzadeh Oskuei S, Zahiri M. A multi-storey DNA nanostructure containing doxorubicin and AS1411 aptamer for targeting breast cancer cells. J Drug Target 2022; 30(10):1106-12. doi: 10.1080/1061186x.2022.2094387 [Crossref] [ Google Scholar]
- Alizadeh F, Yaghoobi E, Imanimoghadam M, Ramezani M, Alibolandi M, Abnous K. Targeted delivery of epirubicin to cancerous cell using copper sulphide nanoparticle coated with polyarginine and 5TR1 aptamer. J Drug Target 2023; 31(9):986-97. doi: 10.1080/1061186x.2023.2274804 [Crossref] [ Google Scholar]
- Moradi E, Zavvar T, Alibolandi M, Ramezani M, Abnous K, Taghdisi SM. Targeted delivery of epirubicin to breast cancer cells using poly-aptamer DNA nanocarriers prepared by the RCA method with multiple repeats of aptamers of FOXM1 and AS1411. Drug Dev Ind Pharm 2023; 49(3):260-70. doi: 10.1080/03639045.2023.2199075 [Crossref] [ Google Scholar]
- Kim M, Lee JS, Kim W, Lee JH, Jun BH, Kim KS. Aptamer-conjugated nano-liposome for immunogenic chemotherapy with reversal of immunosuppression. J Control Release 2022; 348:893-910. doi: 10.1016/j.jconrel.2022.06.039 [Crossref] [ Google Scholar]
- García Melián MF, Moreno M, Cerecetto H, Calzada V. Aptamer-based immunotheranostic strategies. Cancer Biother Radiopharm 2023; 38(4):246-55. doi: 10.1089/cbr.2022.0064 [Crossref] [ Google Scholar]
- Wei Y, Qin G, Wang Z, Zhao C, Ren J, Qu X. Bioorthogonal activation of TLR7 agonists provokes innate immunity to reinforce aptamer-based checkpoint blockade. ACS Nano 2023; 17(6):5808-20. doi: 10.1021/acsnano.2c12313 [Crossref] [ Google Scholar]
- Zheng L, Wu H, Wen N, Zhang Y, Wang Z, Peng X. Aptamer-functionalized nanovaccines: targeting in vivo DC subsets for enhanced antitumor immunity. ACS Appl Mater Interfaces 2023; 15(15):18590-7. doi: 10.1021/acsami.2c20846 [Crossref] [ Google Scholar]
- Zhang J, Li W, Qi Y, Wang G, Li L, Jin Z. PD-L1 aptamer-functionalized metal-organic framework nanoparticles for robust photo-immunotherapy against cancer with enhanced safety. Angew Chem Int Ed Engl 2023; 62(5):e202214750. doi: 10.1002/anie.202214750 [Crossref] [ Google Scholar]
- Zahedipour F, Majeed M, Kesharwani P, Sahebkar A. Cancer immunotherapy via nucleic acid aptamers. In: Kesharwani P, ed. Aptamers Engineered Nanocarriers for Cancer Therapy. Woodhead Publishing; 2023. p. 317-46. 10.1016/b978-0-323-85881-6.00003-8.
- Song W, Hu JJ, Song SJ, Xu Y, Yang H, Yang F. Aptamer-gold nanocage composite for photoactivated immunotherapy. ACS Appl Mater Interfaces 2022; 14(38):42931-9. doi: 10.1021/acsami.2c11089 [Crossref] [ Google Scholar]
- Yang G, Zhang S, Wang Y, Li L, Li Y, Yuan D. Aptamer blocking S-TLR4 interaction selectively inhibits SARS-CoV-2 induced inflammation. Signal Transduct Target Ther 2022; 7(1):120. doi: 10.1038/s41392-022-00968-2 [Crossref] [ Google Scholar]
- Tian L, Shao M, Gong Y, Chao Y, Wei T, Yang K. Albumin-binding lipid-aptamer conjugates for cancer immunoimaging and immunotherapy. Sci China Chem 2022; 65(3):574-83. doi: 10.1007/s11426-021-1168-4 [Crossref] [ Google Scholar]
- Kohlberger M, Gadermaier G. SELEX: critical factors and optimization strategies for successful aptamer selection. Biotechnol Appl Biochem 2022; 69(5):1771-92. doi: 10.1002/bab.2244 [Crossref] [ Google Scholar]
- Qi S, Duan N, Khan IM, Dong X, Zhang Y, Wu S. Strategies to manipulate the performance of aptamers in SELEX, post-SELEX and microenvironment. Biotechnol Adv 2022; 55:107902. doi: 10.1016/j.biotechadv.2021.107902 [Crossref] [ Google Scholar]
- Duan Y, Zhang C, Wang Y, Chen G. Research progress of whole-cell-SELEX selection and the application of cell-targeting aptamer. Mol Biol Rep 2022; 49(8):7979-93. doi: 10.1007/s11033-022-07317-0 [Crossref] [ Google Scholar]
- Mukherjee M, Appaiah P, Sistla S, Bettadaiah BK, Bhatt P. Bio-layer interferometry-based SELEX and label-free detection of patulin using generated aptamer. J Agric Food Chem 2022; 70(20):6239-46. doi: 10.1021/acs.jafc.2c01591 [Crossref] [ Google Scholar]
- Halder S, Thakur A, Keshry SS, Jana P, Karothia D, Das Jana I. SELEX based aptamers with diagnostic and entry inhibitor therapeutic potential for SARS-CoV-2. Sci Rep 2023; 13(1):14560. doi: 10.1038/s41598-023-41885-w [Crossref] [ Google Scholar]
- Parashar A, Bhushan V, Mahanandia NC, Kumar S, Mohanty AK. Non-SELEX method for aptamer selection against β-casomorphin-7 peptide. J Dairy Sci 2022; 105(7):5545-60. doi: 10.3168/jds.2021-21569 [Crossref] [ Google Scholar]
- Razlansari M, Jafarinejad S, Rahdar A, Shirvaliloo M, Arshad R, Fathi-Karkan S. Development and classification of RNA aptamers for therapeutic purposes: an updated review with emphasis on cancer. Mol Cell Biochem 2023; 478(7):1573-98. doi: 10.1007/s11010-022-04614-x [Crossref] [ Google Scholar]
- Cai R, Chen X, Zhang Y, Wang X, Zhou N. Systematic bio-fabrication of aptamers and their applications in engineering biology. Systems Microbiology and Biomanufacturing 2023; 3(2):223-45. doi: 10.1007/s43393-022-00140-5 [Crossref] [ Google Scholar]
- Alkhamis O, Xiao Y. Systematic study of in vitro selection stringency reveals how to enrich high-affinity aptamers. J Am Chem Soc 2023; 145(1):194-206. doi: 10.1021/jacs.2c09522 [Crossref] [ Google Scholar]
- Chan KY, Kinghorn AB, Hollenstein M, Tanner JA. Chemical modifications for a next generation of nucleic acid aptamers. Chembiochem 2022; 23(15):e202200006. doi: 10.1002/cbic.202200006 [Crossref] [ Google Scholar]
- Duan N, Li C, Zhu X, Qi S, Wang Z, Wu S. Simultaneous coupled with Separate SELEX for heterocyclic biogenic amine-specific aptamers screening and their application in establishment of an effective aptasensor. Sens Actuators B Chem 2022; 352(Pt 1):130985. doi: 10.1016/j.snb.2021.130985 [Crossref] [ Google Scholar]
- Yang G, Liu W, Zhao Y, Jiang G, Zhu C, Qu F. Induction of binding sites for RecA aptamers by differentiated-competition capillary Electrophoresis-SELEX. Talanta 2024; 267:125213. doi: 10.1016/j.talanta.2023.125213 [Crossref] [ Google Scholar]
- Poustforoosh A, Faramarz S, Nematollahi MH, Hashemipour H, Negahdaripour M, Pardakhty A. In silico SELEX screening and statistical analysis of newly designed 5mer peptide-aptamers as Bcl-xl inhibitors using the Taguchi method. Comput Biol Med 2022; 146:105632. doi: 10.1016/j.compbiomed.2022.105632 [Crossref] [ Google Scholar]
- Wang Y, Liu X, Wu L, Ding L, Effah CY, Wu Y. Construction and bioapplications of aptamer-based dual recognition strategy. Biosens Bioelectron 2022; 195:113661. doi: 10.1016/j.bios.2021.113661 [Crossref] [ Google Scholar]
- Xiong H, Liu L, Liu X, Jiang H, Wang X. Aptamer-based immune drug systems (AptIDCs) potentiating cancer immunotherapy. Chemistry 2023; 5(3):1656-80. doi: 10.3390/chemistry5030114 [Crossref] [ Google Scholar]
- Yasmeen F, Seo H, Javaid N, Kim MS, Choi S. Therapeutic interventions into innate immune diseases by means of aptamers. Pharmaceutics 2020; 12(10):955. doi: 10.3390/pharmaceutics12100955 [Crossref] [ Google Scholar]
- Thomas BJ, Porciani D, Burke DH. Cancer immunomodulation using bispecific aptamers. Mol Ther Nucleic Acids 2022; 27:894-915. doi: 10.1016/j.omtn.2022.01.008 [Crossref] [ Google Scholar]
- Kamath V. Cancer vaccines: an unkept promise?. Drug Discov Today 2021; 26(6):1347-52. doi: 10.1016/j.drudis.2021.02.006 [Crossref] [ Google Scholar]
- Xu L, Liang J, Wang Y, Ren S, Wu J, Zhou H. Highly selective, aptamer-based, ultrasensitive nanogold colorimetric smartphone readout for detection of CdII. Molecules 2019; 24(15):2745. doi: 10.3390/molecules24152745 [Crossref] [ Google Scholar]
- Yang G, Liu Y, Deng Y, Chen Z, Chen H, Li S. Selection of a high-affinity DNA aptamer for the recognition of cadmium ions. J Biomed Nanotechnol 2021; 17(11):2240-6. doi: 10.1166/jbn.2021.3191 [Crossref] [ Google Scholar]
- Ni S, Zhuo Z, Pan Y, Yu Y, Li F, Liu J. Recent progress in aptamer discoveries and modifications for therapeutic applications. ACS Appl Mater Interfaces 2021; 13(8):9500-19. doi: 10.1021/acsami.0c05750 [Crossref] [ Google Scholar]
- Han J, Gao L, Wang J, Wang J. Application and development of aptamer in cancer: from clinical diagnosis to cancer therapy. J Cancer 2020; 11(23):6902-15. doi: 10.7150/jca.49532 [Crossref] [ Google Scholar]
- Allemailem KS, Almatroudi A, Alsahli MA, Basfar GT, Alrumaihi F, Rahmani AH. Recent advances in understanding oligonucleotide aptamers and their applications as therapeutic agents. 3 Biotech 2020; 10(12):551. doi: 10.1007/s13205-020-02546-1 [Crossref] [ Google Scholar]
- Khan S, Hussain A, Fahimi H, Aliakbari F, Haj Bloukh S, Edis Z. A review on the therapeutic applications of aptamers and aptamer-conjugated nanoparticles in cancer, inflammatory and viral diseases. Arab J Chem 2022; 15(2):103626. doi: 10.1016/j.arabjc.2021.103626 [Crossref] [ Google Scholar]
- Aljohani MM, Cialla-May D, Popp J, Chinnappan R, Al-Kattan K, Zourob M. Aptamers: potential diagnostic and therapeutic agents for blood diseases. Molecules 2022; 27(2):383. doi: 10.3390/molecules27020383 [Crossref] [ Google Scholar]
- Riccardi C, Meyer A, Vasseur JJ, Cavasso D, Russo Krauss I, Paduano L. Design, synthesis and characterization of cyclic NU172 analogues: a biophysical and biological insight. Int J Mol Sci 2020; 21(11):3860. doi: 10.3390/ijms21113860 [Crossref] [ Google Scholar]
- Nikolaeva PA, Moryachkov RV, Raldugina VN, Naumova JO, Novikova TM, Spiridonova VA. Structural analysis of thrombin-binding G-aptamers in presence of bivalent ions. Sib Med Rev. 2022(5):111-3. 10.20333/25000136-2022-5-111-113.
- Xiang W, Peng Y, Zeng H, Yu C, Zhang Q, Liu B. Targeting treatment of bladder cancer using PTK7 aptamer-gemcitabine conjugate. Biomater Res 2022; 26(1):74. doi: 10.1186/s40824-022-00328-9 [Crossref] [ Google Scholar]