Biomed Res Bull. 2(3):103-107.
doi: 10.34172/biomedrb.2024.16
Original Article
Let7i Restoration Inhibits Migration and Proliferation While Inducing Apoptosis in Melanoma Cells by Directly Targeting CXCR4 In Vitro
Babak Sandoghchian Shotorbani 1  , Leila Mahboubi 2, Leila-sadat Hatamnezhad 3, *
, Leila Mahboubi 2, Leila-sadat Hatamnezhad 3, * 
Author information:
1Department of Pediatrics, Oncology Section, Ardabil University of Medical Sciences, Ardabil, Iran
2Department of Pediatrics, Tabriz University of Medical Sciences, Tabriz, Iran
3Department of Dermatology, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
Abstract
Background:
Melanoma is a type of skin cancer that originates from the mutation of melanocytes. miRNAs are small non-coding RNA molecules that bind to their target mRNA and regulate many biological processes. Aberrant expression of microRNAs has been reported in different types of cancer. The present study aimed to determine the role of Let7i in melanoma cancer pathogenesis.
Methods:
In this study, we evaluated the expression level of Let-7 and its potential target genes in specimens obtained from melanoma patients. Then, we transfected the Let-7i mimic and assessed its effect on cell migration, proliferation, and apoptosis using the Scratch test, MTT assay, and Flow cytometry, respectively. Then, the impact of Let7i restoration on chemokine receptor type 4 (CXCR4) expression was assessed by real-time polymerase chain reaction (PCR) and Western blotting. A rescue experiment was performed to determine the association between Let-7 and CXCR4 in melanoma pathogenesis. Moreover, the correlation between Let-7a and CXCR4 expression and clinicopathological features of the patients was evaluated.
Results:
The results indicated that Let-7a was down-regulated, while CXCR4 was up-regulated in specimens derived from melanoma patients. It was also discovered that transfection of Let-7i alleviated the proliferative and metastatic capacity of SK-MEL-3 melanoma cells and led to an increased apoptosis rate in these cells. Rescue experiments demonstrated that Let-7 reduced the metastatic potential of melanoma cells by directly targeting CXCR4. Additionally, a correlation was observed between Let-7 and CXCR4 with lymph node metastasis.
Conclusion:
This study reveals that Let-7 serves as a potential tumor-suppressor in melanoma by directly targeting CXCR4, indicating a possible therapeutic and diagnostic candidate for patients suffering from melanoma.
Keywords: Let-7i, CXCR4, Melanoma, Metastasis, Proliferation, Apoptosis
Copyright and License Information
© 2024 The Author(s).
This is an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Funding Statement
This study was self-funded by the authors and received no external financial support from any funding organization.
Introduction
Melanoma is one of the most dangerous types of skin cancer, primarily caused by UV rays exposure. The accumulation of melanin granules above the keratinocytes is the main cause of melanoma.1-5 MicroRNAs are single-stranded, non-coding RNA molecules regulating various biological processes.6-8 They have been implicated in melanoma progression, survival, and proliferation.9-11 MicroRNAs have been shown to modulate critical regulatory pathways involved in melanoma progression, suggesting their potential role in melanoma treatment.12-18 Let-7i has been identified as a tumor-suppressor microRNA that is down-regulated in gastrointestinal, prostate, and lung cancer, and melanoma.2,6 Let-7i influences cell proliferation, metastasis, and invasion, indicating its potential as a prognostic, diagnostic, and therapeutic tool in cancers.1
Chemokine receptor type 4 (CXCR4) has been identified as a metastasis-promoting gene in cancers such as melanoma. It has been implicated in the progression of cancers by mediating metastasis.10,16 CXCR4, one of the defined oncogenes in cancer, mediates its role through extracellular matrix (ECM) degradation, leading to metastasis and invasion.15,19,20
In this study, we aimed to explore the role of Let7i in melanoma by transfecting its mimic oligonucleotides. We also strived to determine the impact of Let7 up-regulation on proliferation, migration, apoptosis, and the expression of CXCR4 in SK-MEL-3 melanoma cells. Rescue experiments were performed to investigate the exact association between Let-7 and CXCR4 in melanoma pathogenesis. Finally, the correlation between Let-7 and CXCR4 expressions with the clinicopathological features of patients was assessed.
Materials and Methods
Patient Samples
Fifty tissue samples from melanoma patients and 50 from adjacent non-tumor tissues were obtained. All samples were immediately frozen and preserved in liquid nitrogen. The included patients had no history of radiotherapy or chemotherapy.
Cell Culture
SK-MEL-3 cells for this study were obtained from ACCEGEN (Fairfield, New Jersey 07004, USA). The cells were cultured and maintained in a moisture condition containing 5% CO2 at 37 °C. The culture medium comprised Dulbecco’s modified eagle medium (DMEM) supplemented with 10 fetal bovine serum (FBS) + penicillin-streptomycin.
Transfection of microRNA
After seeding cells in 6-well plates, Let7i oligonucleotide mimic (Eurogentec, Belgium) and a negative scramble control were transfected using the Polyplus transfection reagent (France) following the manufacturer’s protocol.
RNA Isolation and Real-Time Polymerase Chain Reaction
Cells were cultured in 6-well plates. RNA was extracted using a TRIzol reagent following the manufacturer’s protocol (PCR Biosystems Ltd, UK). RNAs were reverse-transcribed into cDNAs by a conventional PCR step (PCR Biosystems Ltd, UK) using a cDNA synthesis kit. The ABI QuantStudio 3D Digital PCR System was used to perform the real-time PCR reaction using SYBR Premix Ex Taq (Bio-equip, China.), and the GAPDH gene was used as an internal control.
Western Blotting
First, the proteins were extracted using radioimmunoprecipitation assay (RIPA) lysis buffer (Boster Bio, Pleasanton, CA, USA) and loaded onto sodium dodecyl sulfate (SDS) gels, which were then transferred to polyvinylidene fluoride (PVDF) membranes using a semi-dry transfer system. Membranes were then blocked with 0.5% Tween-20 solved in phosphate-buffered saline (PBS) and incubated for two hours with goat monoclonal antibodies against CXCR and Beta-actin (Boster Bio, Pleasanton, CA, USA). In addition, horseradish peroxidase (HRP)-conjugated rabbit anti-goat secondary antibodies (Boster Bio, Pleasanton, CA, USA) were used for one hour at room temperature. Finally, bands pictured by the western blot imaging instrument were visualized using an enhanced chemiluminescence kit, and the density of bands was qualified using ImageJ software.
As part of the rescue experiment, SK-MEL-3 cell lines were treated with either Let-7 or CXCR4 expression, and CXCR4 expression at the protein level was assessed via Western blotting as described above.
MTT Assay
To assess the viability of the melanoma cell line affected by Let-7i transfection, cells were seeded in 96-well plates, and the cell viability was measured by the MTT test carried out by a specific kit (MyBioSource, Southern California, San Diego, USA) following the manufacturer’s protocol at 570 nm.
In the rescue experiment, SK-MEL-3 cells were treated with either Let-7 or CXCR4 expression to determine the effect of their simultaneous transfection on cell viability.
Wound Healing Assay
In 24-well plates, melanoma cell lines were transfected with the optimal dose of Let7i mimic and scratched with a sterile yellow pipette tip after being filled into the plates and incubated for 48 hours. Finally, a reverse microscope was used to monitor and capture images of cell migration into the scratched area.
Flow Cytometry
When the cells reached 80% confluence, Let7 mimics were transfected. Then, FITC Annexin V Apoptosis Detection (Elabscience, Biomedical Park, Wuhan, China) was used with a flow cytometry instrument (FACSVerseTM – BD Biosciences) according to the manufacturer’s protocol.
Statistical Analysis
GraphPad Prism software version 6.00 was used for statistical analysis based on ANOVA and Student’s t-tests. The chi-square test was used to assess possible correlations between target genes and patients’ clinicopathological features. In this study, a P value of less than 0.05 was considered significant.
Results
Let-7 was down-regulated, while chemokine receptor type 4 was up-regulated melanoma patients
qRT-PCR results showed a reduction in Let-7 expression in tissues obtained from melanoma patients, while CXCR4 expression was elevated (Figure 1).
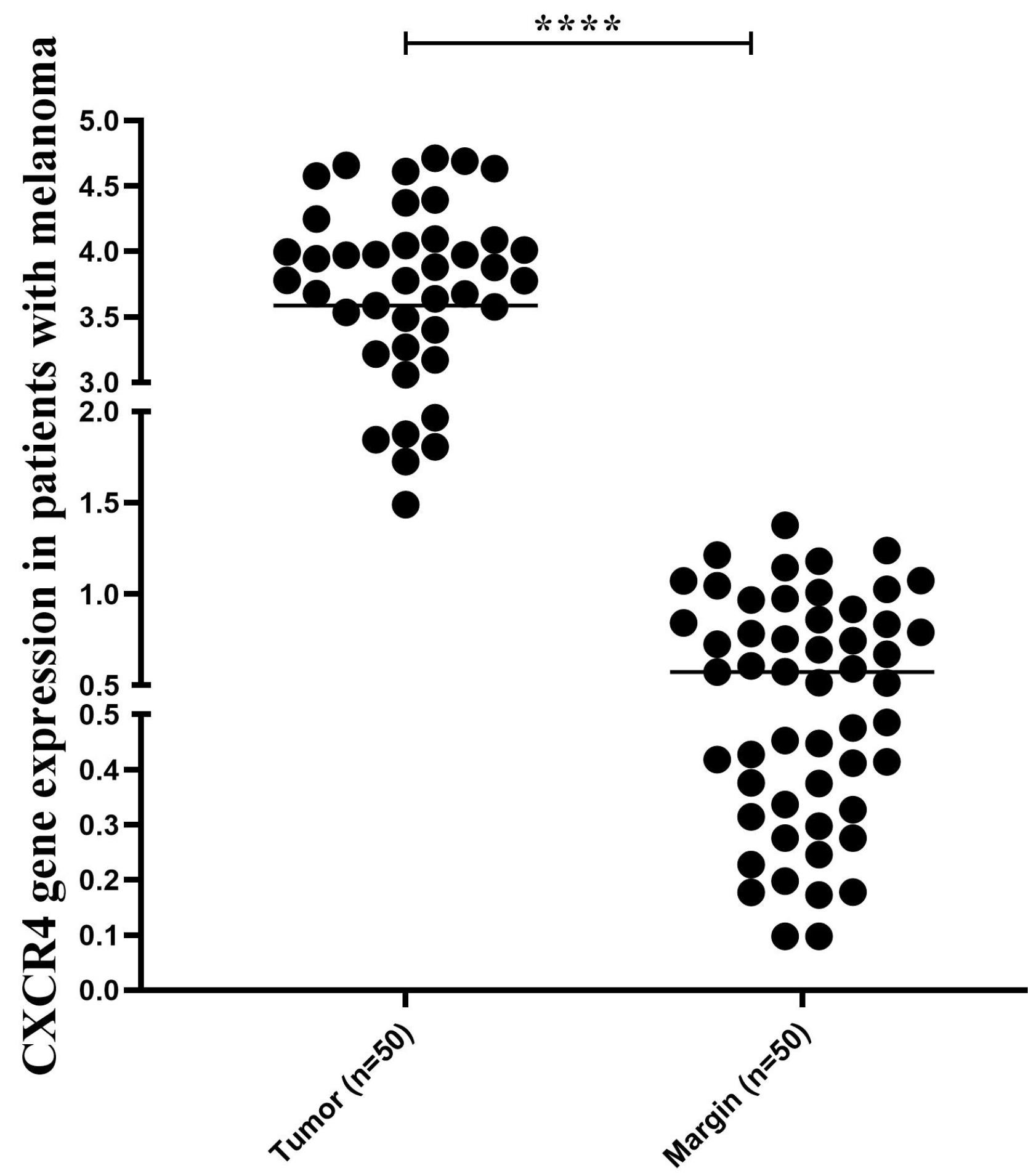
Figure 1.
CXCR4 Gene Expression. Note. CXCR4: Chemokine receptor type 4
.
CXCR4 Gene Expression. Note. CXCR4: Chemokine receptor type 4
Let7 transfection caused up-regulation in melanoma cells
qRT-qPCR was used to measure Let-7 expression in melanoma cells. The results revealed that transfection with 10 nmol (optimum dose) of Let-7 mimics induced up-regulation compared to non-transfected cells (Figure 2).
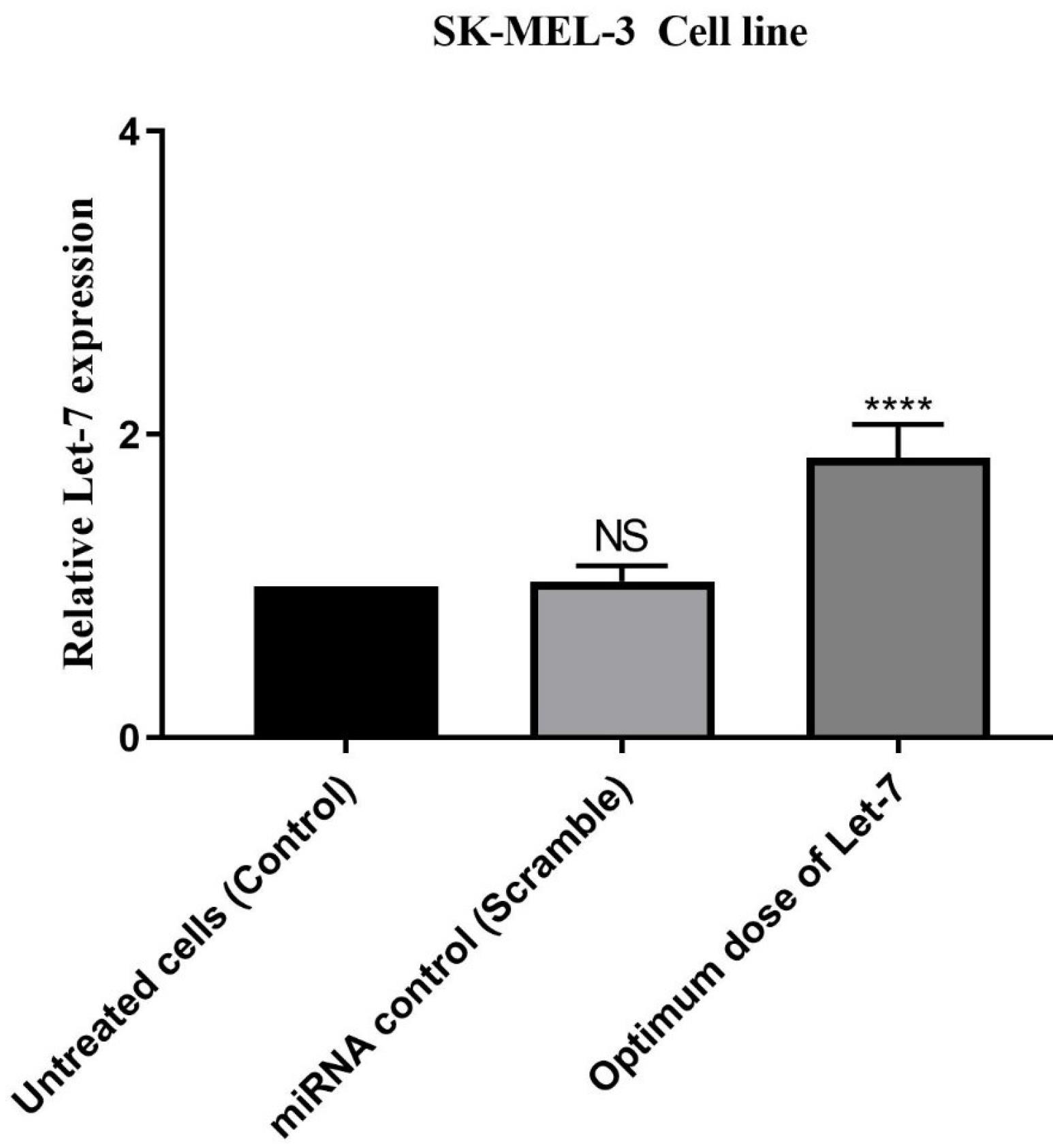
Figure 2.
Let-7 Expression in Optimum Dose
.
Let-7 Expression in Optimum Dose
Let-7 replacement alleviated chemokine receptor type 4 expression
qRT-PCR and western blotting results showed that transfection with Let-7 reduced CXCR4 expression at both the mRNA and protein levels (Figure 3).
Let-7 replacement decreased cell proliferation and increased apoptosis
MTT assay results indicated that Let7i could reduce cell viability in melanoma cells compared to the control group. Flow-cytometry, combined with the MTT assay, demonstrated that Let7i induced apoptosis compared to untreated control cells. Dimethyl sulfoxide (DMSO) was used as a positive control in this study (Figure 4).
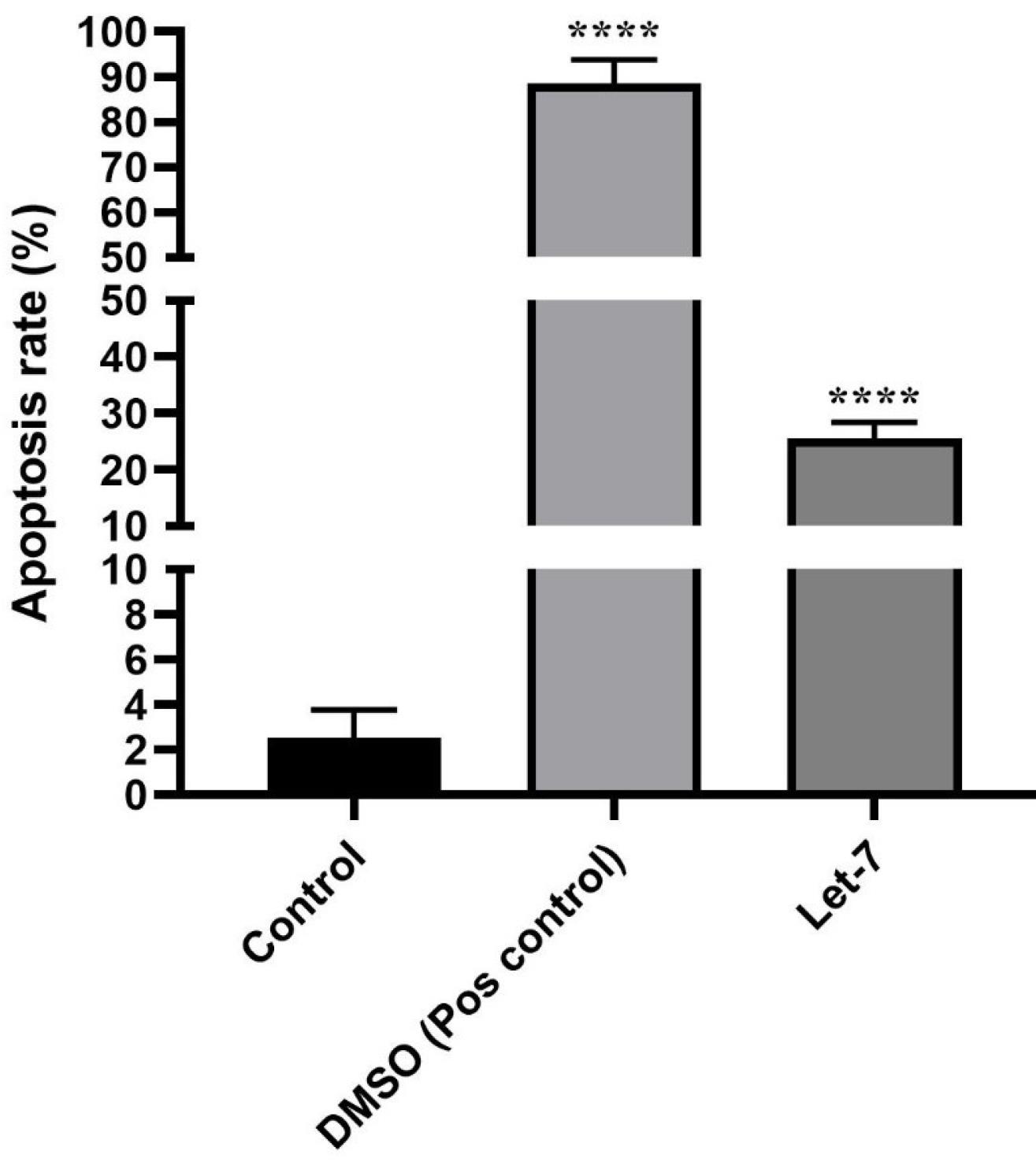
Figure 4.
Apoptosis Rate With Let-7 Compared With DMSO. Note. DMSO: Dimethyl sulfoxide
.
Apoptosis Rate With Let-7 Compared With DMSO. Note. DMSO: Dimethyl sulfoxide
Let-7 replacement reduced migration ability of SK-MEL-3 melanoma cells
Wound healing assay results revealed that Let7 reduced the migration ability of cells compared to unscratched control cells (Figure 5).
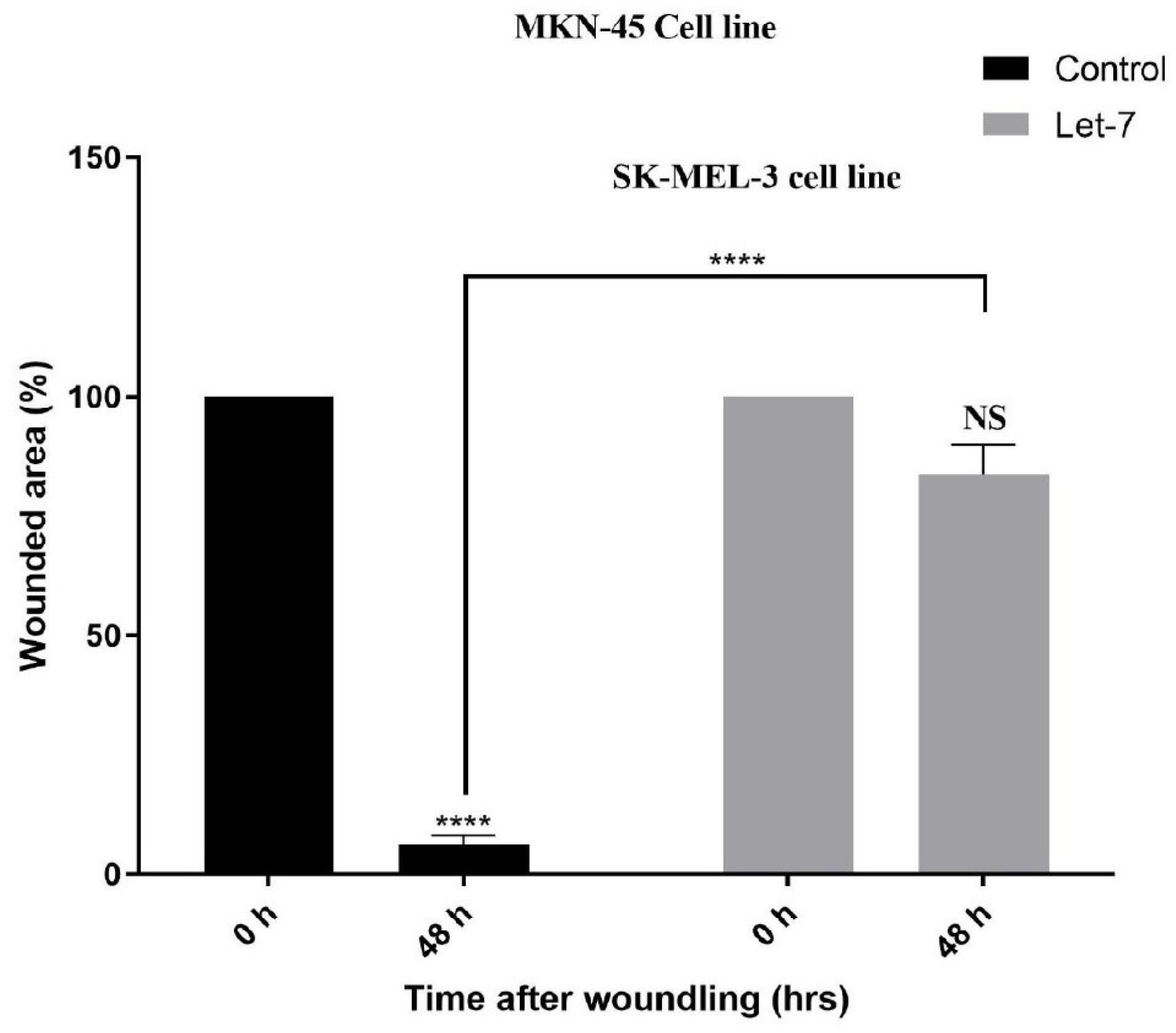
Figure 5.
Wound Healing Assay Result
.
Wound Healing Assay Result
Simultaneous transfection of Let-7 and chemokine receptor type 4 and their effect on chemokine receptor type 4 expression and cell viability
The results obtained from rescue experiments demonstrated that Let-7 transfection reduced CXCR4 expression, while CXCR4 cDNA transfection induced its up-regulation. However, when both Let-7 and CXCR4 cDNA were transfected simultaneously, CXCR4 expression remained untouched. Moreover, MTT assay results showed that Let-7 restoration reduced cell viability, while CXCR4 cDNA transfection increased cell viability. The simultaneous transfection showed a milder effect on cell viability. These results suggest that Let-7 modulates CXCR4 expression (Figure 6).
Let-7 and chemokine receptor type 4 expression correlated with clinicopathological features of patients
The results showed that Let-7 expression was significantly correlated with tumor subtype (P = 0038) and lymph node metastasis (P = 0.014). Regarding CXCR4, an association was observed between tumor size (P = 0.036) and lymph node metastasis (P < 0.0001), as depicted in Figure 7.
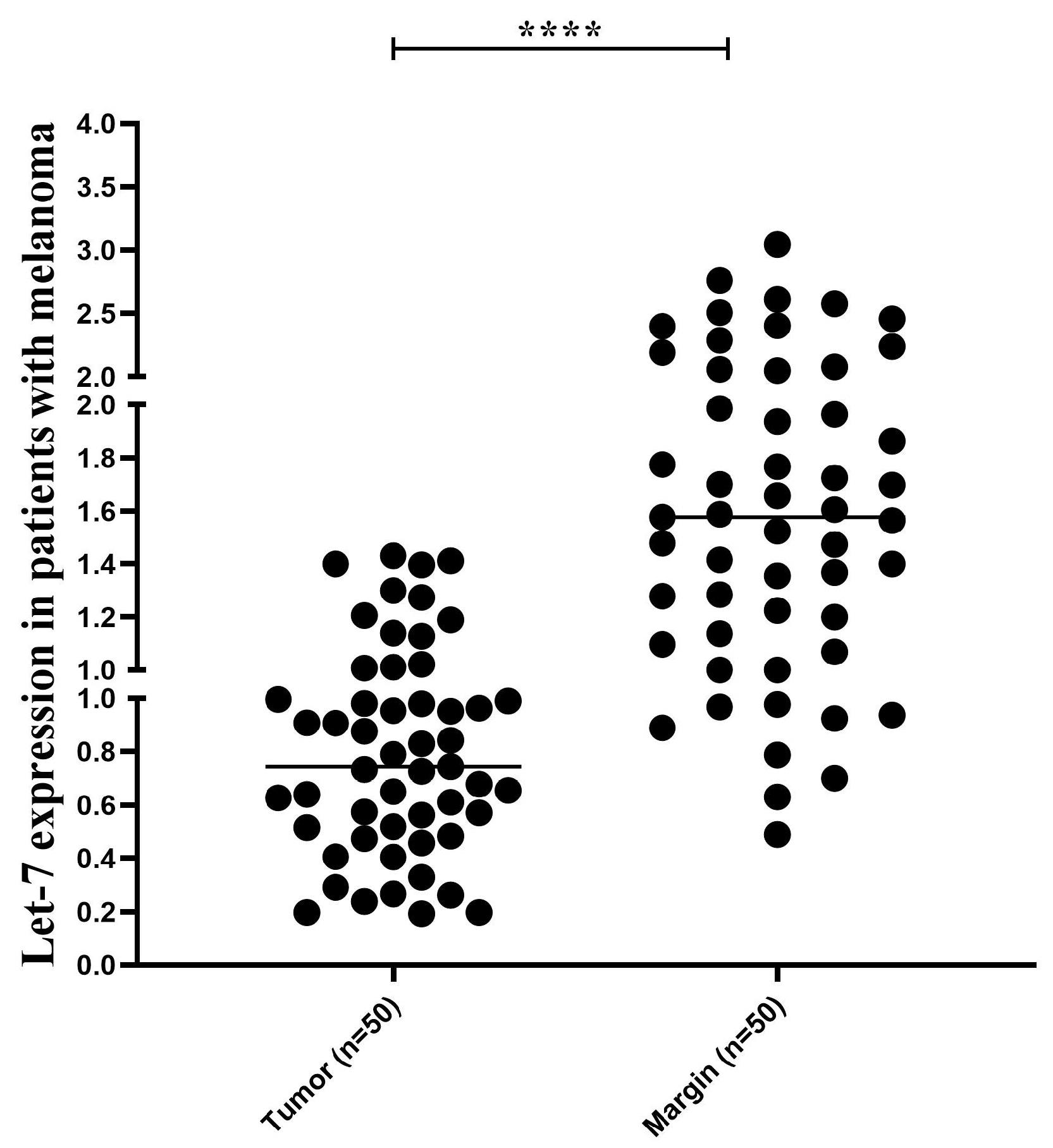
Figure 7.
Let-7 Expression
.
Let-7 Expression
Discussion
Melanoma is a type of skin cancer that spreads rapidly throughout the body. Surgical excision is the main approach for treating primary melanoma; however, melanoma has a high recurrence rate. Consequently, understanding the exact molecular mechanism underlying this aggressive cancer may open a new horizon for its treatment.4
Previous studies have shown that Let-7 reduces cell viability either by decreasing cell proliferation or promoting apoptosis, revealing its role as a potential tumor suppressor in melanoma.12,13 Let 7 up-regulation has also been associated with reduced progression of melanoma.2 A study conducted in 2015 showed that Let-7 transfection reduced the metastatic potential of melanoma cells in vitro.14 Similarly, Let-7 has been implicated in lowering the proliferative capacity of several types of cancers,3,17,21 supporting the findings in our study.
Many genes have been implicated in the pathogenesis of melanoma and are regulated by Let-7. For example, in vitro studies have shown that Let-7 inhibits the differentiation and migration of melanoma cells by directly targeting MMP-9, a gene involved in cell metastasis.7 CXCR4 has also been identified as a potential mediator of melanoma metastasis. In this regard, as a predicted target gene of Let-7, we sought to examine its association with melanoma carcinogenesis. Our findings suggested that restoring Let-7i expression can reduce melanoma cell metastasis and proliferation by directly targeting CXCR4 at both the mRNA and protein levels.9,19
Conclusion
In conclusion, this study showed that Let-7 serves as a potential tumor-modulating microRNA in melanoma carcinogenesis, reducing melanoma cell viability and metastasis in vitro. Moreover, CXCR4 was found to be up-regulated in melanoma patients. Furthermore, Let-77 restoration reduced CXCR4 expression, a critical gene involved in melanoma carcinogenesis and metastatic pathways. Moreover, our findings suggest that Let-7 mediates its anti-cancer effects by directly inhibiting CXCR4. To sum up, both Let-7 and CXCR4 may serve as potential therapeutic targets in melanoma pathogenesis. However, further studies are needed to determine their precise roles in melanoma pathogenesis.
Authors’ Contribution
Conceptualization: Leila-sadat Hatamnezhad.
Data curation: Babak Sandoghchian.
Formal analysis: Babak Sandoghchian.
Investigation: Babak Sandoghchian.
Methodology: Babak Sandoghchian.
Project administration: Leila Mahboobi.
Resources: Babak Sandoghchian.
Software: Babak Sandoghchian.
Supervision: Leila-Sadat Hatamnezhad.
Validation: leila Mahboobi.
Visualization: Leila-Sadat Hatamnezhad.
Writing–original draft: leila Mahboobi.
Writing–review & editing: Leila-Sadat Hatamnezhad.
Competing Interests
None.
Ethical Approval
This study was approved by Research Ethics Committees of Vice-Chancellor in Research Affairs - Tabriz University of Medical Sciences (Ethical code: IR.TBZMED.VCR.REC.1398.164)
References
- Golabi Aghdam S, Ebrazeh M, Hemmatzadeh M, Seyfizadeh N, Gowhari Shabgah A, Azizi G. The role of microRNAs in prostate cancer migration, invasion, and metastasis. J Cell Physiol 2019; 234(7):9927-42. doi: 10.1002/jcp.27948 [Crossref] [ Google Scholar]
- Alkafaji HA, Raji A, Rahman HS, Zekiy AO, Adili A, Jalili M. Up-regulation of KISS1 as a novel target of Let-7i in melanoma serves as a potential suppressor of migration and proliferation in vitro. J Cell Mol Med 2021; 25(14):6864-73. doi: 10.1111/jcmm.16695 [Crossref] [ Google Scholar]
- Chirshev E, Oberg KC, Ioffe YJ, Unternaehrer JJ. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin Transl Med 2019; 8(1):24. doi: 10.1186/s40169-019-0240-y [Crossref] [ Google Scholar]
- Davis LE, Shalin SC, Tackett AJ. Current state of melanoma diagnosis and treatment. Cancer Biol Ther 2019; 20(11):1366-79. doi: 10.1080/15384047.2019.1640032 [Crossref] [ Google Scholar]
- Domingues B, Lopes JM, Soares P, Pópulo H. Melanoma treatment in review. Immunotargets Ther 2018; 7:35-49. doi: 10.2147/itt.S134842 [Crossref] [ Google Scholar]
- Fattore L, Costantini S, Malpicci D, Ruggiero CF, Ascierto PA, Croce CM. MicroRNAs in melanoma development and resistance to target therapy. Oncotarget 2017; 8(13):22262-78. doi: 10.18632/oncotarget.14763 [Crossref] [ Google Scholar]
- Fu TY, Chang CC, Lin CT, Lai CH, Peng SY, Ko YJ. Let-7b-mediated suppression of basigin expression and metastasis in mouse melanoma cells. Exp Cell Res 2011; 317(4):445-51. doi: 10.1016/j.yexcr.2010.11.004 [Crossref] [ Google Scholar]
- Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol 2019; 20(1):21-37. doi: 10.1038/s41580-018-0045-7 [Crossref] [ Google Scholar]
- Kim J, Mori T, Chen SL, Amersi FF, Martinez SR, Kuo C. Chemokine receptor CXCR4 expression in patients with melanoma and colorectal cancer liver metastases and the association with disease outcome. Ann Surg 2006; 244(1):113-20. doi: 10.1097/01.sla.0000217690.65909.9c [Crossref] [ Google Scholar]
- Lu J, Luo H, Liu X, Peng Y, Zhang B, Wang L. miR-9 targets CXCR4 and functions as a potential tumor suppressor in nasopharyngeal carcinoma. Carcinogenesis 2014; 35(3):554-63. doi: 10.1093/carcin/bgt354 [Crossref] [ Google Scholar]
- Mannavola F, Tucci M, Felici C, Stucci S, Silvestris F. miRNAs in melanoma: a defined role in tumor progression and metastasis. Expert Rev Clin Immunol 2016; 12(1):79-89. doi: 10.1586/1744666x.2016.1100965 [Crossref] [ Google Scholar]
- Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res 2008; 18(5):549-57. doi: 10.1038/cr.2008.45 [Crossref] [ Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000; 403(6772):901-6. doi: 10.1038/35002607 [Crossref] [ Google Scholar]
- Serguienko A, Grad I, Wennerstrøm AB, Meza-Zepeda LA, Thiede B, Stratford EW. Metabolic reprogramming of metastatic breast cancer and melanoma by let-7a microRNA. Oncotarget 2015; 6(4):2451-65. doi: 10.18632/oncotarget.3235 [Crossref] [ Google Scholar]
- Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer 2016; 15:18. doi: 10.1186/s12943-016-0502-x [Crossref] [ Google Scholar]
- Sowder ME, Johnson RW. Bone as a preferential site for metastasis. JBMR Plus 2019; 3(3):e10126. doi: 10.1002/jbm4.10126 [Crossref] [ Google Scholar]
- Thammaiah CK, Jayaram S. Role of let-7 family microRNA in breast cancer. Noncoding RNA Res 2016; 1(1):77-82. doi: 10.1016/j.ncrna.2016.10.003 [Crossref] [ Google Scholar]
- Aftab MN, Dinger ME, Perera RJ. The role of microRNAs and long non-coding RNAs in the pathology, diagnosis, and management of melanoma. Arch Biochem Biophys 2014; 563:60-70. doi: 10.1016/j.abb.2014.07.022 [Crossref] [ Google Scholar]
- Thyagarajan A, Shaban A, Sahu RP. MicroRNA-directed cancer therapies: implications in melanoma intervention. J Pharmacol Exp Ther 2018; 364(1):1-12. doi: 10.1124/jpet.117.242636 [Crossref] [ Google Scholar]
- Wang CA, Tsai SJ. Regulation of lymphangiogenesis by extracellular vesicles in cancer metastasis. Exp Biol Med (Maywood) 2021; 246(19):2048-56. doi: 10.1177/15353702211021022 [Crossref] [ Google Scholar]
- Wang X, Cao L, Wang Y, Wang X, Liu N, You Y. Regulation of let-7 and its target oncogenes. Oncol Lett 2012; 3(5):955-60. doi: 10.3892/ol.2012.609 [Crossref] [ Google Scholar]