Biomed Res Bull. 1(4):130-134.
doi: 10.34172/biomedrb.2023.25
Original Article
Evaluation of the Effect of MiR-206 on the Expression of Toll-like Receptor 2 and TLR4 in the Melanoma A-375 Cells
Mojtaba Gholinezhad 1  , Bahareh Sartipi 1, Babak Sandoghchian Shotorbani 1, *
, Bahareh Sartipi 1, Babak Sandoghchian Shotorbani 1, * 
Author information:
1Department of Immunology Group, Tabriz University of Medical Science, Tabriz, Iran
Abstract
Background:
Melanoma is one of the most deadly skin cancers in humanity, which imposes extensive costs on governments. Fortunately, the death rate has decreased despite the increasing incidence of melanoma. Various treatment methods, including surgery, chemotherapy, radiotherapy, targeted therapy, and the like, are used to treat this disease. However, despite recent advances in diagnosis and treatment, melanoma remains one of the leading causes of death in the world. One of the new treatment methods is based on microRNAs, one of which is miRNA 206. The aim of this study was to determine the effect of MIR-206 on Toll-like receptor 2 (TLR2) and TLR4 in the melanoma cell line (A-375).
Methods:
The present in vitro study was performed on melanoma cancer cells in RPMI640 culture medium containing 10% fetal calf serum at 37 ° C and 5% CO2. After counting and dividing the cells, the cells inside the plate were stimulated in over time with the MIR-206 protein. Incubation was performed. After 48 hours, the cells were collected, the total mRNA was extracted, and the expression levels of TLR2 and TLR4 were examined by real-time polymerase chain reaction.
Results:
The results demonstrated that the expression of TLR2 and TLR4 IL6 decreased in the melanoma cell line (A-375) in exposure to MIR-206. The results also showed the expression of nuclear factor kappa B in the melanoma cell line.
Conclusion:
The findings confirmed the role of MIR-206 as a tumor suppressor in melanoma cancer. These results suggest that MIR-206 may be a new treatment for melanoma in the near future. Accordingly, MIR-206 can be considered a potential target in the gene therapy of patients with melanoma cancer, and one of the advantages of MIR-206 gene therapy in melanoma cancer over chemotherapy is the reduction of cytotoxicity and its invasiveness.
Keywords: MIR-206, TLR2, TLR4, Melanoma
Copyright and License Information
© 2024 The Author(s).
This is an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
The incidence of skin cancers is higher than that of all other cancers. Among them, melanoma is the deadliest and is increasing. The incidence rate doubles every decade. The death rate has decreased compared to the past and has reached about 11%.1 Melanoma causes about 9 000 deaths and costs $3.5 billion annually. On average, a person in the United States loses 20.4 years of potential due to melanoma, compared to 16.6 years for other malignant cancers.2 Melanoma is a multifactorial disease in which two factors play a role, namely, genetic predisposition and exposure to sunlight, especially ultraviolet exposure.1 About 5%‒12% of melanomas are hereditary, which usually have different mutations than non-hereditary ones.3 The most common genetic mutation known in hereditary melanomas is CDKN2A, which is also found in spontaneous cases.4 Melanoma is divided into nodular, superficial spreading, and lentigo maligna categories. Other types include acral and mucosal melanoma, and sunlight plays no role in their creation.5 A biopsy is used to diagnose melanoma, and Clark and Breslow criteria are utilized to evaluate it. Various treatment methods have been reported, among them are surgery, chemotherapy, and targeted therapy, each of which has its own advantages and disadvantages.6 Recent studies have indicated the important role of microRNAs (miRNAs) in the development of melanoma. MiRNAs are short (about 22 nucleotides) non-coding RNAs that regulate gene expression post-transcriptionally by binding to the 3’-UTR or 5’-UTR of mRNA. They act in two general ways to target, namely, by targeting mRNA degradation and/or inhibiting mRNA translation. A large body of research has so far been performed to clarify melanoma-specific dysregulated miRNAs.7 miRNAs are transcribed by the polymerase II enzyme, and about 1,900 miRNAs have been identified in humans. MiRNAs are synthesized in the nucleus as pri-miRNAs and then exported to the cytoplasm for the rest of the processing steps to take place there. One type of miRNA can target several distinct mRNAs, and a single mRNA can be processed by several types of miRNAs.8 MiRNAs regulate the progression of tumors through their roles in migration, invasion, cell proliferation, angiogenesis, and resistance to apoptosis. Thus, different miRNAs play a role in cancer cells by decreasing or increasing their expression.9 Toll-like receptors (TLRs) are part of type I integral membrane glycoproteins that play an important role in host defense by recognizing a wide range of pathogens. To date, 10 functional TLRs have been recognized in humans. TLRs are expressed on the surface of various immune cells. They are recognized as one of the main components of inflammation, infections, and several immune-related diseases. Most TLRs, including TLRs 2 and 4, use MyD88 as an adapter protein to activate the nuclear factor kappa B pathway and release inflammatory factors. TLR4 can also use the Toll/interleukin-1 receptor-domain-containing adapter-inducing interferon-β adapter.10 It has recently been identified that TLRs are expressed not only on immune cells but also on various cancer cells, including melanoma.11 TLRs can have different roles in cancers; some of them have a tumorigenesis effect, while others have a tumor suppressor effect. In fact, the effects of TLRs depend on the type of cancer.
Materials and Methods
Ethical Considerations
The A-375 cell line was purchased in vial form from the Pasteur Institute of Iran. This is a basic science, laboratory-based study, and there is no intervention in the living organism, so there is no need to obtain consent. This study was approved by the regional ethics committee with ethics code ID IR.TBZMED.VCR.REC.1401.031. Considering that the existing study is at the cellular level, the sample size is not necessary.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism software, and a one-way ANOVA test was used for the statistical comparison of gene expression levels. A P value was used for all relationships, and P < 0.05 was considered statistically significant.
Cell Culture
Melanoma cancer cell lines A-375 and C-6181 were purchased from the Pasteur Cell Bank. The reason for choosing these cell lines was the high prevalence of malignancy and the relatively fewer studies on this line. After counting, one million cells were transferred to a flask and cultured in RPMI 1640 culture medium + 10% fetal bovine serum (FBS) in an amount of 3–4 mL in an incubator with CO2 at 37 °C. According to growth conditions, previous similar studies, and the morphology of the studied cells, the best culture medium considered for the melanoma cancer cell line A-375 is the RPMI 1640 culture medium, which is supplemented with fetal calf serum at 10% and 20% and provides highly suitable environmental conditions for the growth of A-375 melanoma cancer cells. RPMI 1640 was developed by RPMI in Buffalo, New York. Serum protects cells from physical damage in culture media. FBS is rich in growth factors and has less immunoglobulin than other serums. Nonetheless, the addition of 10% serum to the culture medium eventually adds 4.8 mg of additional protein per mL of the culture medium and makes it difficult to extract and process the original and final product. It was checked for the absence of contamination, and the necessary cell passage was performed accordingly. The cells were frozen when they reached the logarithmic phase of growth, and the required amount of cell stock was prepared. The indicated experiments were performed when the cells reached the desired density. Then, 100 units/mL of penicillin to prevent the growth of gram-positive bacteria, 100 µg/mL of streptomycin to prevent the growth of gram-negative bacteria, 50 µg/mL of gentamicin to prevent the growth of gram-positive and gram-negative bacteria, as well as mycoplasma, kanamycin, and the antifungal drug amphotericin were used to provide antibiotics in the culture medium.
Freezing and Storing Cell Lines
First, the cells were trypsinized and counted, then the prepared cryo-freezing medium was added to the cell sediment and transferred to the cryotube and then to the COOL BOX rack and a -85 freezer. Cryopreservation solution is a mixture of culture medium, FBS, and preservatives, which are combined in a certain ratio and added to the cells. The amount of FBS was 900 μL, and dimethyl sulfoxide was 100 μL for a 1 mL freezing series.
RNA Extraction
The cells were cultured in 6-well plates. After 48 hours of treatment with MIR-206 as the time period of the cells, the supernatant of the plates was separated and centrifuged in separate Falcons for 10 minutes at a speed of 1300 rpm. Then, the supernatant solution was removed and 1000 µL of TRIzol was added to it, and the solutions separated in the previous step were returned to the corresponding well. Next, 1000 µL from each well was transferred to 2 ml DNAse. RNA -free microtubes, and 300 µL of chloroform were added to each tube and shaken well. Separated phases were observed after staying at -20 degrees for 10 minutes. The tubes were centrifuged at 12 000 rpm for 15 minutes at 4 °C, and 3 distinct phases were observed. The upper phase, which consists of RNA, was separated with the RNAase free sampling head and transferred to another microtube without RNAase free, and 1.5 times its volume (700 μL) of isopropanol was added to it, shaken, and kept for 15 minutes at -20 °C. Subsequently, it was centrifuged for 20 minutes at 1200 rpm at 4 °C. After centrifugation, a white precipitate was observed at the end of each microtube. The supernatant was decanted, and 1000 μL of 75% alcohol was added to each tube and centrifuged again at 7500 rpm for 5 minutes at 4 °C. After centrifugation, the excess alcohol was emptied, then 300 µL DEPC water was added to the tube and placed in a 55-degree heater for 10 minutes, and the RNA sample was prepared for cDNA synthesis. To store the RNA sample for a short time, the microtube was transferred to an -80-degree freezer.
Determining the Concentration of Extracted RNA
About 10 µL of dd water was placed on the nanodrop device, and the lens was washed. In addition, 1.5 µL of RNA were placed on the nanodrop device. The amount of protein and phenol contamination was recorded, and different concentrations of RNA were obtained. To compare them, it is necessary that their concentrations be similar. According to the lowest concentration, all samples were compared and multiplied by a number between 1 and 5. The number chosen should be such that the product does not exceed 12 (since the final volume of the mixture of water and RNA was 12 µL). The frozen RNA was brought to -4 °C, and the tubes were kept on ice during all steps.
cDNA Synthesis
After normalizing the RNAs, the indicated volume of water was added to each microtube, and then 1 µL of random hexamer primer was added to each tube. Next, to bind the primers to single-stranded RNA structures, the above mixture was incubated for 5 minutes at 60 °C in a thermocycler. After the incubation, 2 µL dNTP, 0.5 µL RT (reverse transcriptase), and 4.5 µL of buffer, prepared as stock, were added to the tubes to reach a final volume of 20 µL. After a short spin, the tubes were transferred to the thermocycler, and the cDNAs were synthesized according to the programming settings of the device. This polymerase chain reaction (a device made by Bio-Rad Company) was performed with the temperature conditions in Table 1 and Figure 1. Then, the synthesized cDNAs were transferred to -20 °C for short-term storage and -80 °C for long-term storage.
Table 1.
Specific Primers for HMGA1, HMGB1, and β-actin
|
Primer
|
|
Sequence
|
| β-actin |
F |
5´- CAAGATCATCACCAATGCCT - 3´ |
| R |
5´ - CCCATCACGCCACAGTTTCC - 3´ |
| MIR-206 |
F |
5´- TTTCAAACCGCACGGATA - 3´ |
| R |
5´- GACCTTAAGAATCTATGG - 3´ |
| TLR2 |
F |
5´- CCCTAATGTTAAGAGGTA - 3´ |
| R |
5´- CAAATAATTCGGGATAT - 3´ |
| TLR4 |
F |
5´- AATCACAATTGGATCCTA - 3´ |
| R |
5´- TTGGTCAGTCCCCCAAAC - 3´ |
Note. HMGA1: High-mobility group A1; HMGB1: High-mobility group protein B1.
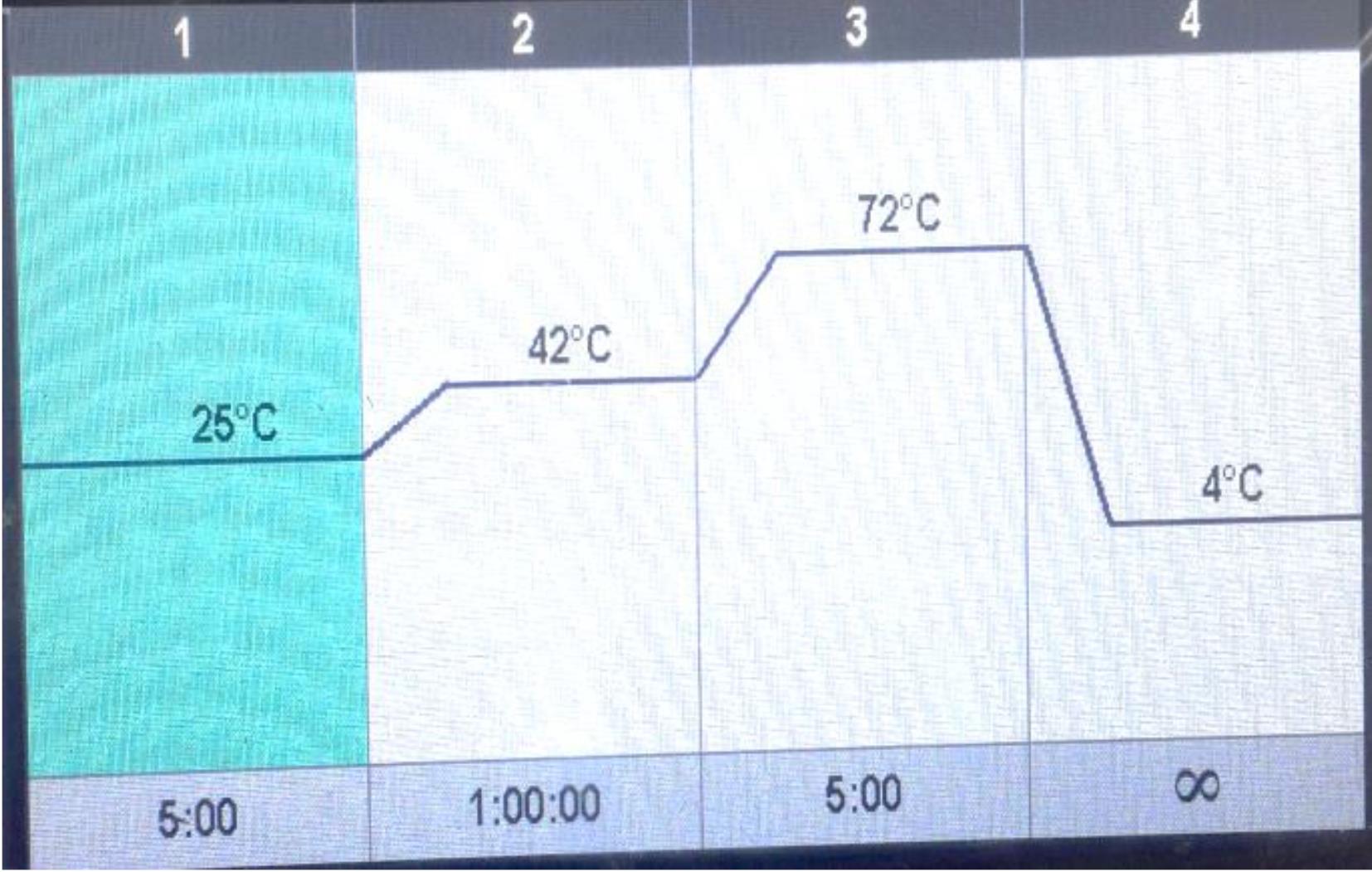
Figure 1.
Temperature Conditions
.
Temperature Conditions
Real-time Polymerase Chain Reaction
First, specific amounts of Master, H20, primers, and cDNA were transferred to the micro-tube, and after a short rotation, they were placed in the real-time polymerase chain reaction (RT-PCR) machine with specific temperature conditions (Table 1). Next, the remaining procedures were performed according to the protocol of Green and Sambrook.12
Measurement of Cytotoxicity
Overall, 2000 cells were cultured in each well of small 96-well plates, and the final volume of each well reached 200 µL using RPMI and was kept overnight in an incubator containing 5% CO2. Then, the liquid from each well was removed, and different concentrations of MIR-206 were added to it (2, 4, 6, and 8 mg/mL). For each concentration, three wells were allocated, and 5 wells were examined as controls without drugs. The cells were incubated with different amounts of MIR-206 at different times (24, 48, 72, and 96 hours). After the end of the incubation period, the3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide (MTT) test was performed by preparing a 5 mg/mL solution of MTT yellow powder (Sigma Company) and then adding 20 µL of it to the wells of the microplate, and finally, the plates were incubated for another 4 hours. During this time, the MTT dye was absorbed by the mitochondria and turned into purple crystals, then the liquid in each well was removed, and 100 µL of acidic isopropanol was added to dissolve the crystals and calorimetry. After a few minutes, the crystals at the bottom of the well were completely dissolved. The optical absorbance of each well was read at a wavelength of 540 nm with an enzyme-linked immunosorbent assay device.
Results
The TLR2 and TLR4 expressions were downregulated by MIR-206 in the A-375 cell line. To determine whether the expression of TLR2 and TLR4 in A-375 cells was regulated by MIR-206, the cells were treated at 42°C for 1 hour and returned to 37 °C at the indicated times. The expression of TLR2 reached its lowest value around 12 and 24 hours after heat shock. TLR4 expression was also regulated within 3 hours, and TLR4 expression began to decrease after 9 hours (Figure 2).
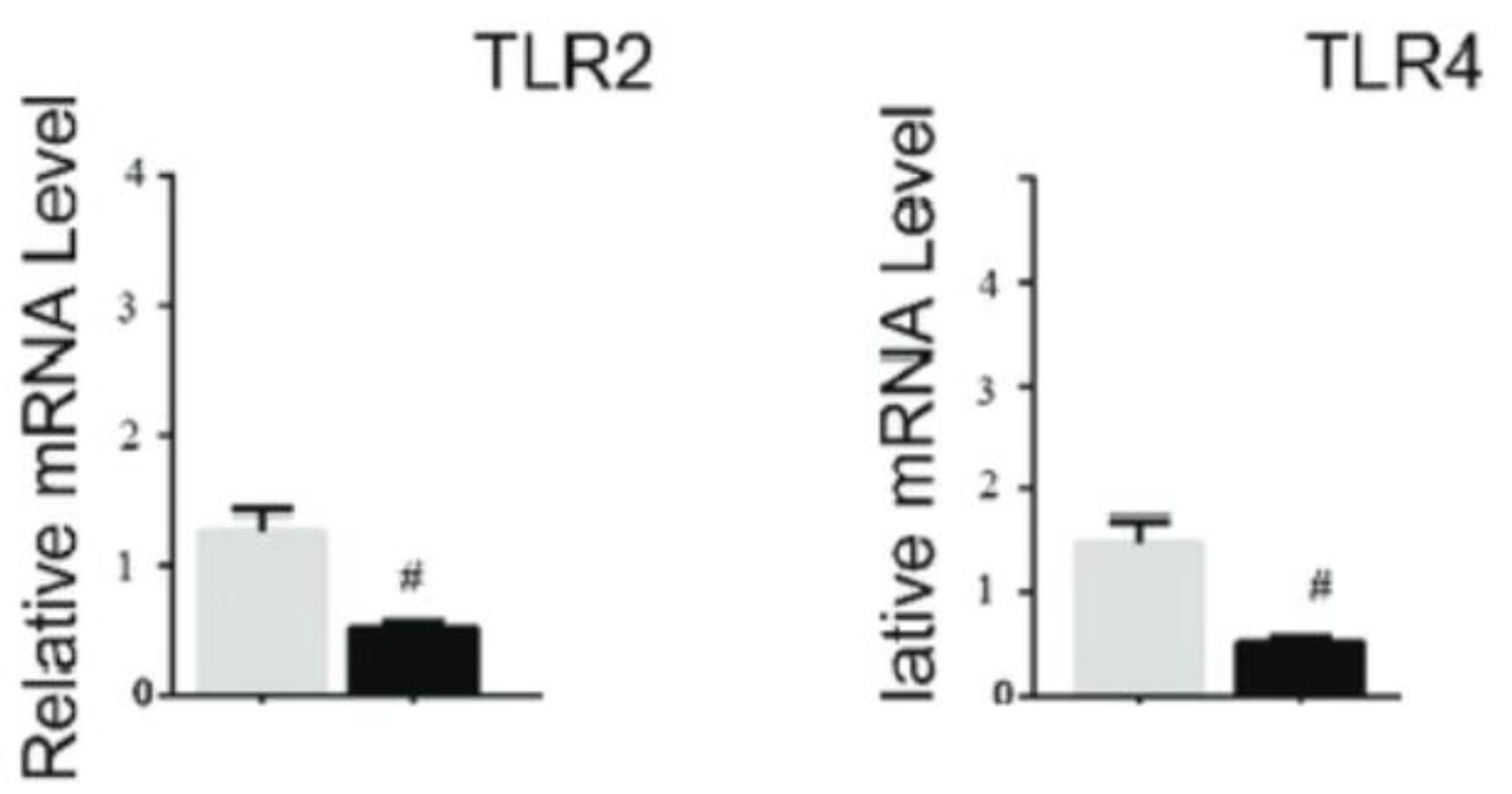
Figure 2.
Expression of TLR2 and TLR4 After Receiving MIR-206. Note. TLR2: Toll-like receptor 2; TLR4: Toll-like receptor 4
.
Expression of TLR2 and TLR4 After Receiving MIR-206. Note. TLR2: Toll-like receptor 2; TLR4: Toll-like receptor 4
3-[4,5-Dimethylthiazol-2-yl]-2,5 Diphenyl Tetrazolium Bromide Assay
Given that the effect of MIR-206 is concentration- and time-dependent, to investigate this property at different times with various concentrations and to ascertain how long each concentration leads to this property, melanoma cell death was compared at different times. The statistical analysis showed a significant difference (P < 0.001, Figure 3).
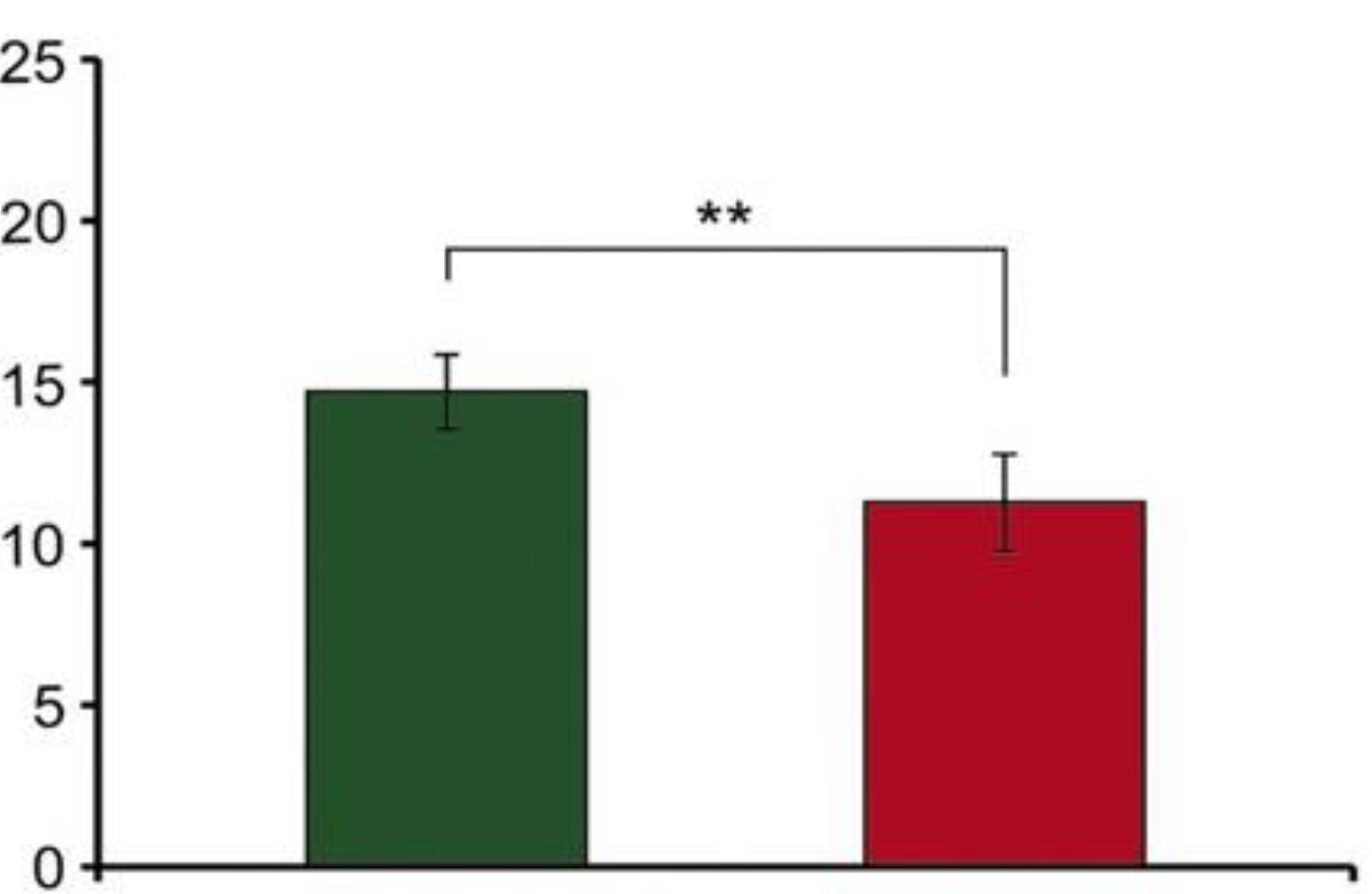
Figure 3.
Inhibitory Effects of MIR-206
.
Inhibitory Effects of MIR-206
Discussion
Melanoma is responsible for about 1% of all skin malignancies. In general, women demonstrate a better prognosis and survival than men, possibly due to the stronger interaction of sex hormones with melanoma cells and the immune system than men, but the exact mechanism remains unclear.13 The evidence obtained based on new studies indicates that the time course of fatal melanomas varies significantly according to the thickness of the melanoma at the time of diagnosis. For melanomas less than 1.0 mm, most deaths occur between 5 and 20 years after diagnosis, while for thicker melanomas, the opposite is true, with most deaths occurring within the first 5 years.14 Melanoma arises from the malignant transformation of melanocytes, cells scattered throughout the body that synthesize melanin, a light-protective pigment. Melanoma can arise from pigment-producing cells in the eyes, digestive system, reproductive system, sinuses, meninges, and especially the skin.15,16 Melanoma is usually observed in old age, when the accumulation of mutations in melanocytes, either inherited or acquired over time, leads to the transformation of melanocytes into melanoma. The average age of melanoma diagnosis in the United States is 65 years, and the average age of death is 71 years.17,18 Surgery is one of the primary treatments for early stages of melanoma, and after complete removal of the tumor, radiotherapy can be used as an adjuvant treatment. Due to the large number of metastases, low accessibility and difficulty in detecting small metastatic lesions, surgical treatment of metastatic melanoma is highly unlikely.19,20 One of the new treatment modalities is targeted therapy that inhibits the mutated BRAF and MEK proteins. However, recent research has shown that continuous treatment with these agents fails due to the development of genetic mutations that cause disease resistance to treatment.21 MiRNAs are a group of short, non-coding RNAs with a length of 21‒22 nucleotides. These transcripts participate in the silencing of target genes after transcription. MiRNAs perform their function by binding to the 3’UTR portion of the gene. They participate in various processes, including cell growth, development, and differentiation. Their disorder causes various pathologies, including cancer. MiRNA 206 is part of the miRNA family and plays an important role in the growth and development of muscle cells.22 MiRNAs have been shown to essentially contribute to melanoma pathogenesis. The indicated role is through the regulation of the microphthalmia-associated transcription factor, which is a key regulator in the differentiation, proliferation, and survival of melanocytes and melanomagenesis.23 It has been demonstrated that miRNAs perform the same function as oncogenes or anti-oncogenes, target genes involved in tumorigenesis and melanoma, and play a vital role in immunotherapy. Moreover, evidence indicates that some miRNAs can have opposite effects on different tumors24. Cai et al concluded that decreased levels of miRNA 206 worsened the prognosis and distant metastases of osteosarcoma patients. Likewise, the overexpression of miRNA 206 reduced the ability of cancer cells to proliferate and invade.25 It has been found in various studies that miRNA 206 is disturbed in lung, colon, cervix, and breast cancers, considering that it is involved in apoptosis, proliferation, migration, invasion, angiogenesis, and drug resistance in lung cancer cells.26 In a study by Robert et al, microRNA-206 induced G1 arrest in melanoma by inhibiting CDK4 and Cyclin D and reduced the growth and invasion of multiple melanoma cell lines. This paper supports miR-206 as a tumor suppressor in melanoma,27 which is consistent with the results of our study. Tian et al revealed that the serum level of miRNA 206 was significantly lower in melanoma patients compared to healthy subjects.28 In a study conducted by DiVincenzo et al, the downregulation of miR-206 and other related miRNAs was detected using MiRNA Human NanoString and the mRNA 360 signaling assay to measure the expression of miRNAs.29
Conclusion
The results of this study confirmed the role of MIR-206 as a tumor suppressor in melanoma cells, which is associated with the reduction of metastatic ability and proliferation capacity, as well as the induction of apoptosis in cancer cells in vitro. These results indicate that MIR-206 may be a novel treatment for melanoma in the near future. However, more research is needed for replacement therapy as a new treatment method. It can be concluded that MIR-206 can be considered a potential target in gene therapy for melanoma patients, and one of the advantages of MIR-206 gene therapy in melanoma compared to chemotherapy is the reduction of cytotoxic effects and its low invasiveness.
Authors’ Contribution
Conceptualization: Babak Sandoghchian Shotorbani.
Data curation: Mojtaba Gholinezhad.
Formal analysis:Bahareh Sartipi.
Funding acquisition: Babak Sandoghchian Shotorbani.
Investigation: Bahareh Sartipi.
Methodology: Bahareh Sartipi.
Project administration: Mojtaba Gholinezhad.
Resources:Mojtaba Gholinezhad.
Software: Mojtaba Gholinezhad.
Supervision: Babak Sandoghchian Shotorbani.
Validation: Babak Sandoghchian Shotorbani.
Visualization: Babak Sandoghchian Shotorbani.
Writing–original draft: Mojtaba Gholinezhad.
Writing–review & editing: Babak Sandoghchian Shotorbani.
Competing Interests
The authors declare no conflict of interests.
Funding
None.
References
- Dzwierzynski WW. Melanoma risk factors and prevention. Clin Plast Surg 2021; 48(4):543-50. doi: 10.1016/j.cps.2021.05.001 [Crossref] [ Google Scholar]
- Ekwueme DU, Guy GP Jr, Li C, Rim SH, Parelkar P, Chen SC. The health burden and economic costs of cutaneous melanoma mortality by race/ethnicity-United States, 2000 to 2006. J Am Acad Dermatol 2011; 65(5 Suppl 1):S133-43. doi: 10.1016/j.jaad.2011.04.036 [Crossref] [ Google Scholar]
- Rebecca VW, Sondak VK, Smalley KS. A brief history of melanoma: from mummies to mutations. Melanoma Res 2012; 22(2):114-22. doi: 10.1097/CMR.0b013e328351fa4d [Crossref] [ Google Scholar]
- Mehnert JM, Kluger HM. Driver mutations in melanoma: lessons learned from bench-to-bedside studies. Curr Oncol Rep 2012; 14(5):449-57. doi: 10.1007/s11912-012-0249-5 [Crossref] [ Google Scholar]
- Bobos M. Histopathologic classification and prognostic factors of melanoma: a 2021 update. Ital J Dermatol Venerol 2021; 156(3):300-21. doi: 10.23736/s2784-8671.21.06958-3 [Crossref] [ Google Scholar]
- Davis LE, Shalin SC, Tackett AJ. Current state of melanoma diagnosis and treatment. Cancer Biol Ther 2019; 20(11):1366-79. doi: 10.1080/15384047.2019.1640032 [Crossref] [ Google Scholar]
- Bennett PE, Bemis L, Norris DA, Shellman YG. miR in melanoma development: miRNAs and acquired hallmarks of cancer in melanoma. Physiol Genomics 2013; 45(22):1049-59. doi: 10.1152/physiolgenomics.00116.2013 [Crossref] [ Google Scholar]
- Weidle UH, Ausländer S, Brinkmann U. Micro RNAs promoting growth and metastasis in preclinical in vivo models of subcutaneous melanoma. Cancer Genomics Proteomics 2020; 17(6):651-67. doi: 10.21873/cgp.20221 [Crossref] [ Google Scholar]
- Latchana N, Ganju A, Howard JH, Carson WE 3rd. MicroRNA dysregulation in melanoma. Surg Oncol 2016; 25(3):184-9. doi: 10.1016/j.suronc.2016.05.017 [Crossref] [ Google Scholar]
- Cui J, Chen Y, Wang HY, Wang RF. Mechanisms and pathways of innate immune activation and regulation in health and cancer. Hum Vaccin Immunother 2014; 10(11):3270-85. doi: 10.4161/21645515.2014.979640 [Crossref] [ Google Scholar]
- Yang Y, Feng R, Wang YZ, Sun HW, Zou QM, Li HB. Toll-like receptors: triggers of regulated cell death and promising targets for cancer therapy. Immunol Lett 2020; 223:1-9. doi: 10.1016/j.imlet.2020.04.002 [Crossref] [ Google Scholar]
- Green MR, Sambrook J. Quantification of DNA by real-time polymerase chain reaction (PCR). CSH Protoc 2018; 2018(10):pdb-rot095034. doi: 10.1101/pdb.prot095034 [Crossref] [ Google Scholar]
- Eddy K, Chen S. Overcoming immune evasion in melanoma. Int J Mol Sci 2020; 21(23):8984. doi: 10.3390/ijms21238984 [Crossref] [ Google Scholar]
- Baade PD, Whiteman DC, Janda M, Cust AE, Neale RE, Smithers BM. Long-term deaths from melanoma according to tumor thickness at diagnosis. Int J Cancer 2020; 147(5):1391-6. doi: 10.1002/ijc.32930 [Crossref] [ Google Scholar]
- Lo JA, Fisher DE. The melanoma revolution: from UV carcinogenesis to a new era in therapeutics. Science 2014; 346(6212):945-9. doi: 10.1126/science.1253735 [Crossref] [ Google Scholar]
- Jenkins RW, Fisher DE. Treatment of advanced melanoma in 2020 and beyond. J Invest Dermatol 2021; 141(1):23-31. doi: 10.1016/j.jid.2020.03.943 [Crossref] [ Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020; 70(1):7-30. doi: 10.3322/caac.21590 [Crossref] [ Google Scholar]
- Matthews NH, Li WQ, Qureshi AA, Weinstock MA, Cho E. Epidemiology of Melanoma. In: Cutaneous Melanoma: Etiology and Therapy [Internet]. Brisbane, AU: Codon Publications; 2017. p. 3-22. 10.15586/codon.cutaneousmelanoma.2017.ch1.
- Strojan P. Role of radiotherapy in melanoma management. Radiol Oncol 2010; 44(1):1-12. doi: 10.2478/v10019-010-0008-x [Crossref] [ Google Scholar]
- Amaria RN, Reddy SM, Tawbi HA, Davies MA, Ross MI, Glitza IC. Neoadjuvant immune checkpoint blockade in high-risk resectable melanoma. Nat Med 2018; 24(11):1649-54. doi: 10.1038/s41591-018-0197-1 [Crossref] [ Google Scholar]
- Kakadia S, Yarlagadda N, Awad R, Kundranda M, Niu J, Naraev B. Mechanisms of resistance to BRAF and MEK inhibitors and clinical update of US Food and Drug Administration-approved targeted therapy in advanced melanoma. Onco Targets Ther 2018; 11:7095-107. doi: 10.2147/ott.s182721 [Crossref] [ Google Scholar]
- Khalilian S, Hosseini Imani SZ, Ghafouri-Fard S. Emerging roles and mechanisms of miR-206 in human disorders: a comprehensive review. Cancer Cell Int 2022; 22(1):412. doi: 10.1186/s12935-022-02833-2 [Crossref] [ Google Scholar]
- Simmons JL, Pierce CJ, Al-Ejeh F, Boyle GM. MITF and BRN2 contribute to metastatic growth after dissemination of melanoma. Sci Rep 2017; 7(1):10909. doi: 10.1038/s41598-017-11366-y [Crossref] [ Google Scholar]
- Varrone F, Caputo E. The miRNAs role in melanoma and in its resistance to therapy. Int J Mol Sci 2020;21(3). 10.3390/ijms21030878.
- Cai WT, Guan P, Lin MX, Fu B, Wu B, Wu J. MiRNA-206 suppresses the metastasis of osteosarcoma via targeting Notch3. J Biol Regul Homeost Agents 2020; 34(3):775-83. doi: 10.23812/20-72-a-26 [Crossref] [ Google Scholar]
- Pan JY, Sun CC, Bi ZY, Chen ZL, Li SJ, Li QQ. miR-206/133b cluster: a weapon against lung cancer?. Mol Ther Nucleic Acids 2017; 8:442-9. doi: 10.1016/j.omtn.2017.06.002 [Crossref] [ Google Scholar]
- Georgantas RW 3rd, Streicher K, Luo X, Greenlees L, Zhu W, Liu Z. MicroRNA-206 induces G1 arrest in melanoma by inhibition of CDK4 and cyclin D. Pigment Cell Melanoma Res 2014; 27(2):275-86. doi: 10.1111/pcmr.12200 [Crossref] [ Google Scholar]
- Tian R, Liu T, Qiao L, Gao M, Li J. Decreased serum microRNA-206 level predicts unfavorable prognosis in patients with melanoma. Int J Clin Exp Pathol 2015; 8(3):3097-103. [ Google Scholar]
- DiVincenzo MJ, Schwarz E, Ren C, Barricklow Z, Moufawad M, Yu L, et al. Expression patterns of microRNAs and associated target genes in ulcerated primary cutaneous melanoma. J Invest Dermatol 2023;143(4):630-8.e3. 10.1016/j.jid.2022.09.654.